NFAT (Nuclear Factor of activated T Cells) family is composed by five members: NFATC (1, 2, 3 and 4) and NFAT5. They play a crucial role in the transcription of cytokines and other genes critical in immune response.
Genes encoding NFAT proteins are transcribed in almost every tissue, although the expression of individual proteins is more tissue restricted. NFAT1, NFAT2 and NFAT4 are expressed in immune cells, but NFAT2 has been implicated in cardiac valve development and NFAT3 in cardiac hypertrophy.
NFATC are regulated at several levels. When intracellular calcium rises, the calmodulin-dependent serine-threonine phosphatase calcineurin dephosphorylates NFAT proteins in the cytoplasm, exposing their NLS (Nuclear Localization Sequence) and inducing their translocation to the nucleus. The complete pathway involved is shown here.
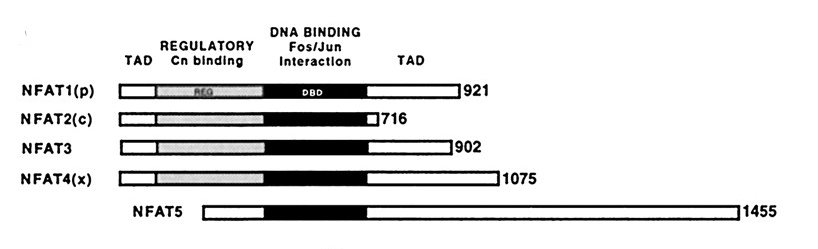
All NFATC can be divided in 3 domains:
The DBD contributes also to the NFAT regulation forming cooperative and stable DNA-binding complexes with dimers of the AP-1 family (Fos / Jun) at composite NFAT:AP-1 DNA elements that have been identified close in NFAT-regulated genes. This combinatorial mechanism results in integration of the calcium/calcineurin and protein kinase C/mitogen-activated protein kinase signaling pathways, which are activated in immune and non-immune processes.
On the other hand, NFAT5 lacks the regulatory domain: it does not cooperate with Fos/Jun and it is a nuclear phosphoprotein, not being unphosphorylated by calcineurin. Spite of these differences, its sequence homology classifies it as a NFAT: it has a high sequence identity in is DBD (up to 43%) with the other NFATs, and its consensus DNA binding site closely resembles the NFAT one.
The respectively consensus sites sequences are:
NFATC: GGAAAATT
NFAT5: TGGAAANNYNY
The Rel-like domain owes its name to its similarity to the Rel domains found in NFkB family of transcription factors. If we look into the structure of the DNA-protein contacts, the DNA recognition loop of NFAT5 (RARYLTEG) contains and arginine rather than a histidine at position 3, and thus is an intermediate between Rel (RFRYxCEG) and NFATC (RAHYxTEG).
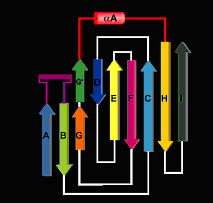
Respect to their secondary structure, the core structural motif of the NFATc DBD consists of a 10 stranded antiparallel beta-barrel assembled by the packing of 3 beta-sheets. The two primary sheets form the core of the beta-barrel, with the third sheet capping off one end and without contacting DNA. One prominent feature is the loops in the DBD, specially bA-bA and bG'-bH This last loop also adapts an alpha-helix conformation.

Bases in the core NFAT site (GGAAAA) and the AP-1 site (TGTTTCA) interact with NFAT and Fos-Jun, respectively, in the major groove, and the intervening spacer sequence (TT) interacts with an NFAT arginine residue in the minor groove. In the NFAT recognition of DNA the AB loop and the alpha-helical segment of the linker, similar to the NFAT5 one, supply side chains that contact bases in the major groove. The AB loop is the homologue of the "recognition loop" in NF-B p50 and its conformation and contacts closely resemble those of its p50 counterpart.
To explain this site specific recognition, there are some important aminoacids and DNA atoms involved. Arg 430 and Arg 421 participate in bidentate hydrogen-bond interactions with O6 and N7 of Gua 1 and Gua 2, respectively. The guanidinium groups of the arginines stack upon each other, following the stack of the corresponding guanine bases. In NFAT, the side-chain amide group of Gln 571 extends this stack and engages in bidentate hydrogen-bonding with Ade 3. The GGA sequence thus recognized seems to be the essential element of an NFAT site. The remaining A:T base pairs in the GGAAAA sequence, which appears to be well conserved among many NFAT sites, are specified by van der Waals contacts with their thymine methyl groups. The ring of Tyr 424 is the partner for thymines 3' and 4'; the aliphatic part of Arg 572 is the partner for thymines 5' and 6'. Both residues are also anchored by hydrogen bonds to the DNA backbone.
Another important loop is the E'F one, which bridges across the DNA backbone from the major to the minor groove. Arg 537, at the end of this loop, dips into the minor groove and donates a hydrogen bond to the carbonyl of Thy 8For a deeper explanation of this topics, look the brief explanations of what has been published since now.