The Rel/NF-kB transcription factors constitute one of the most important families of regulatory transcription factors. Members of this family are essential for diverse biological functions such as the regulation of innate and adaptive immunity, development, and apoptosis in a wide array of eukaryotes from Drosophila to man.
Like most transcription factors, dimers of NF-kB proteins modulate transcription by directly binding to enhancer sequences located in the regulatory regions of numerous genes. These DNA sequences are collectively known as kB DNA sequences. In mammals, the Rel/NF-kB dimers arise from five polypeptides, p50, p52, p65, c-Rel, and RelB. The most abundant of these dimers are the p50/p65 heterodimer (which plays a more elaborate role than other factors in regulating gene expression) and the p50 homodimer. The existence of some, but not all, of the other possible dimers have been shown to exist in cells. The pathway involved in their activation is shown here.

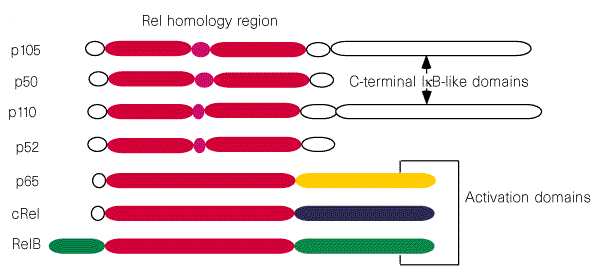
As we can see in the former figure, the NF-kB family can be divided into two subgroups on the presence or absence of an activation domain. p50 and p52 do not contain a distinct activation domain and belong to class I. The other three members constitute the class II sub-family. It is generally believed that the homodimers of p50 and p52 and the p50/p52 heterodimer function as transcriptional repressors. The remaining combinations of dimeric NF-kB proteins, containing at least one monomer of p65, c-Rel, or RelB, act as activators.
Rel/NF-kB proteins share a region that shows over 45% sequence similarity across the entire family. This region, known as the Rel homology region (RHR) is responsible for DNA binding and subunit dimerization. As expected, Rel/NF-kB proteins also share similar structures. Most of the RHR is folded into two immunoglobulin-like domains connected by a 10-amino acid linker; the N-terminal domain confers sequence specificity in DNA binding, it has ~240 residues building a 9 stranded beta-sandwich, and a pair of anti-parallel helices is inserted between strands 5 and 6, although these helices are not conserved in all RHR proteins. The C-terminal domain is involved in dimerization as well as DNA backbone recognition, it is formed by ~100 residue domain containing a 7-stranded beta-sandwich. Strands 2, 3 and 4 on one side of the sandwich form a dimer interface by packing against equivalent strands in the other subunit.
These structures show that, unlike most other transcription factors, NF-kB dimers do not use any secondary structure for contacting DNA. All the DNA-contacting residues emanate from loops connecting secondary structures. Crystal structures of these complexes suggest that in their free form the N-terminal domains should be flexible with respect to the dimerization domain.
Secondary structures of the heterodimer subunits are equivalent, apart from a 32 aminoacids insert in the N-terminal domain of p50 that adds a second alpha-helix. Dimerization in the heterodimer is localized in the C-terminal domains and consists of a hydrophobic core stapled by various polar interactions. The backbones of the dimerization domains are highly similar and one relevant aspect is the hydrogen bond between homologous residues Asp254 of p50 and Asn200 of p65, which is unique in the heterodimer. In the homodimers an interaction between homologous residues is energetically unfavourable because it this juxtaposes two like charges. Also of interest are nine polar contacts, most of which occur between the backbones of hydrophobic core residues and the side chains of hydrogen-bonding residues. These results are consistent with the observation that the affinity between homodimers of p50 and p65 is weaker than for the heterodimers.
As basically the contacts between the homodimer and DNA are equivalent to the ones between the heterodimer and it, we are going to explain the overall contacts in which are involved p50/p65 heterodimers and DNA.
Base-specific binding by p50 to the subsite occurs through the residues Arg54, Arg56, Tyr57, Glu60, His64 and Lys241. The strict conservation of the first three guanines in the p50 subsite is determined by the hydrogen bonds made by His64 with G5, and by Arg56 and Arg54 with G4 and G3. This interaction is strengthened by Glu60, which hydrogen-bonds with these arginines and makes base-specific contacts to C5 and C4.
Binding to the 3' subsite by the p65 subunit occurs through Arg33, Arg35, Tyr36, Glu39 and Arg187. The base conservation observed in B DNA sequences at positions +3 and +4 is a result of the hydrogen bonds formed with Arg33 and Arg35. Glu39 strengthens this conservation by hydrogen-bonding to the N4 groups of C+3 and C+4. Unlike its p50 counterpart Lys241, the geometry of Arg 187 is limited by interactions with Glu39. Another characteristic is that both p50 and p65 make extensive contacts with the ribose phosphate backbone of DNA.
The p50/p65 heterodimer has a particularly high DNA affinity compared to most eukaryotic transcription factors. This affinity cannot be accounted for by direct DNA readout alone as other transcription factors make similar numbers of contacts. Three other sources might contribute to this high binding affinity. First, interdomain interactions in both subunits might provide a high degree of cooperativity between the two DNA-contacting domains. Second, although the target DNA consists of two subsites, phosphate contacts beyond subsite limits could provide coupling energy in the protein-DNA binding pathway. Finally, the extensive dimer interface could also generate large cooperativity in DNA binding.
For a deeper explanation of this topics, look the brief explanations of what has been published since now.