BIANA
BIANA: Biologic Interaction and Network Analysis
Biological Interactions and Network Analysis Using BIANA Cytoscape Plugin
Emre Güney & Javier García García
November 3, 2009
Contents
- 1. Introduction
- 2. Installation
- 3. Usage
- 4. BIANA administration commands
- 4.1 Executing administration commands
- 4.2 Create a new BIANA database
- 4.3 Populate an existing BIANA Database
- 4.4 Drop an existing BIANA Database
- 4.5 Create a new unification protocol in a BIANA Database
- 4.6 Drop an unification protocol in a BIANA Database
- 5. BIANA working commands
- 5.1 Preliminary introduction
- 5.2 Start a working session
- 5.3 User Entity Sets: Characteristics and methods
- 5.3.1 Create a new set of user entities
- 5.3.2 Duplicate a user entity set
- 5.3.3 Remove a user entity set
- 5.3.4 Select nodes in a user entity set
- 5.3.5 Clear previous selection of nodes in a user entity set
- 5.3.6 Tag selected nodes in a user entity set
- 5.3.7 Delete selected nodes in a user entity set
- 5.3.8 Create a sub user entity set
- 5.3.9 Intersection of user entity sets
- 5.3.10 Union of user entity sets
- 5.3.11 View and Export
- 5.3.12 Creating a network in a User Entity Set
- 5.3.13 Network Randomization
- 5.3.14 Output network of relations
- 5.3.15 Show relation details
- 6. BIANA External Databases Parsers
- 6.1 Available External Databases Parsers
- 6.2 Preparing my data to use the generic parser
- 6.3 Creating your own parser for your own data
- 6.4 Command line arguments accepted by parsers
- 6.5 Attributes and types recognized by BIANA and defining new ones
- 6.6 Proposed unification protocol
- 7. Additional administration utilities
- 8. Glossary
- 9. Frequently Asked Questions (FAQs)
1. Introduction
1.1 BIANA Overview
BIANA (Biologic Interactions and Network Analysis) is a biological database integration and network management framework written in Python. BIANA is a Python framework designed to achieve two major goals: i) the integration of multiple sources of biological information, including biological entities and their relationships, and ii) the management of biological information as a network where entities are nodes and relationships are edges.
BIANA uses a generic method to find entries of a given molecule that are equivalent across different biological data repositories. Moreover, BIANA incorporates and empowers a variety of network analysis methods through NetworkX python package. In addition to integrating all major biological repositories, BIANA is easily adaptable to newly created data repositories. The BIANA framework is an extension of the Protein Interaction and Network Analysis (PIANA), which was focused on protein-protein interactions. BIANA bridges the network visualization of Cytoscape and the network analysis capabilities of NetworkX with customizable data integration for any type of relationships between genes and their products. BIANA address the challenge of unambiguously gathering all available data for the biological entities of interest and working with their networks.
The main focus of BIANA is biological database unification and to let user decide how to do it. In order to make sure that BIANA would be freely accessible by anybody, BIANA framework uses either free open-source software (Python, MySQL, MySQLdb, NetworkX, Cytoscape, CD-HIT) or publicly available free software (BLAST). For users who want to skip these software requirements, we provide BIANA web server at the price of loosing freedom on how to decide data unification, relinquishing to incorporate user-defined data and obliging primitive network analysis & visualization.
1.2 BIANA Architecture
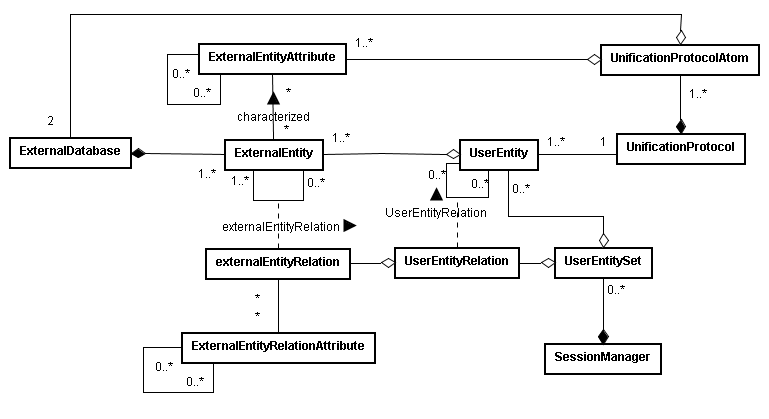
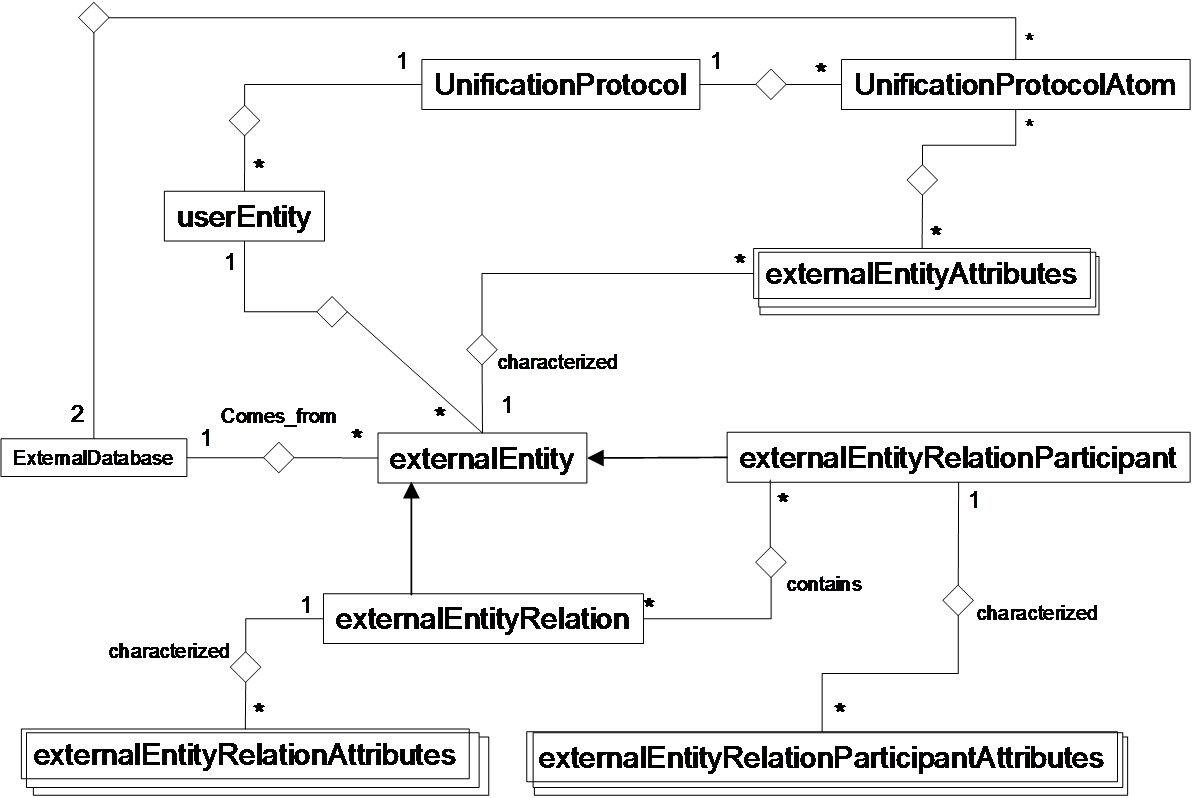
BIANA uses a high level abstraction schema to define databases providing any kind of biological information (both individual entries and their relationships) (See Figure 1.1 and 1.2). Any data source that contains biologic or chemical data parsed by BIANA is defined as an external database. Similarly BIANA integration approach adopts the concept of external entity, corresponding to entries in external databases. For example, a Uniprot entry (a protein), a GenBank entry (a gene), an IntAct interaction (an interaction), a KEGG pathway (a metabolic relation) or a PFAM alignment are all represented as external entities.
In order to achieve data uniformity, in the cases where the data repository supplies relations, both participants and relation itself are considered as external entities. The relation itself is annotated as external entity relation (a subtype of external entity). External entity objects are characterized by several attributes, such as database identifiers, sequence, taxonomy, description or function. Each external entity relation object is further characterized by some attributes like detection method and reliability. Alternatively, the participants in external entity relations can have their particular attributes like role and cardinality.
BIANA unifies external data inserted into its database using its parsers based on a specific protocol. This protocol, called unification protocol, consists of a set of rules that determine how data in various data sources are combined (crossed). Each rule is composed of attributes that have to be crossed and the external databases which are going to be used. The set of external entities that are decided to be "equivalent" with respect to a given unification protocol is called user entity. User entities inherit all the attributes of their included external entries. Thus, BIANA utilizes user entries specific to a certain unification protocol chosen by the user. User can either use provided built-in unification protocols or create his/her own unification protocols. As an example, a user may be interested in creating a unification protocol defined by crossing similar sequences and same taxonomy between two or more databases and crossing entities by uniprot accession code. The advantages of this integration approach are: 1) BIANA database only contains raw data (with exactly the same nomenclature and identifiers of the original data source), therefore does not entail any assumption on data integration allowing user to specify how the integration should be done; 2) User can use information from a single database or the combination of multiple databases, selecting which ones he wants to use at each moment; 3) User can know exactly how was the original data, and do a backtracking of his/her integration approach.
1.3 BIANA Data Unification Approach
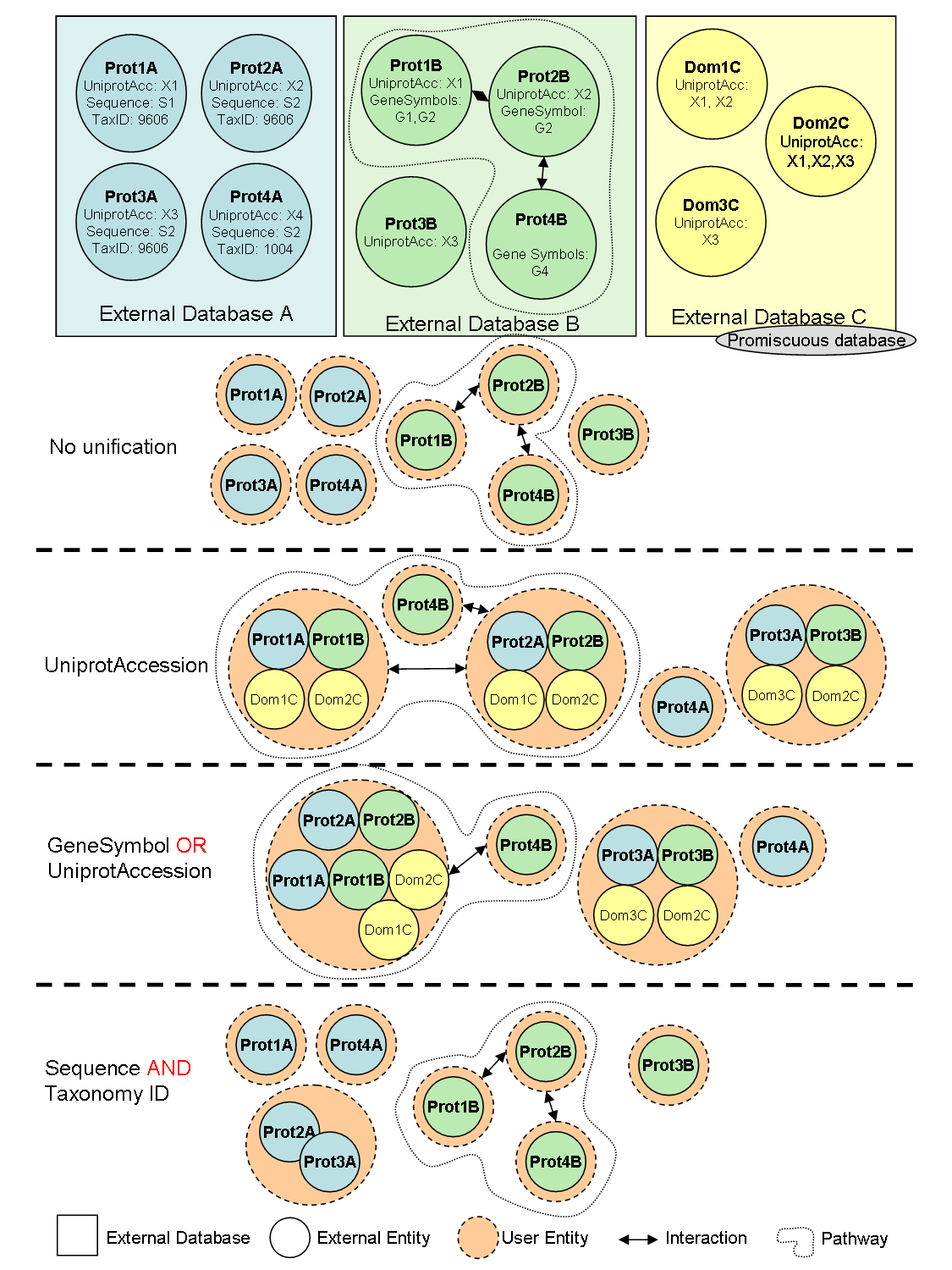
The integration protocol is defined by the user deciding which type of common features will be used on the equivalence of database entries (i.e. by using sequence identity, matching identifiers or sharing domain). Equivalent entries in distinct biological database sources are represented as a single node in a network, while their relationships with other nodes are considered edges. The main advantage of BIANA over other unification software is being user driven. By default, all entities (usually molecules) coming from different databases are considered non-equivalent. Then user decides which databases are unified and which attributes are used to consider molecules as equivalent. This approach has the advantage that the user decides the databases (including his/her own data if any) and identifiers being used. On the other hand, rather inconvenient than a drawback, it is the responsibility of the user to know which attributes are provided in each database.
BIANA also offers insertion of an External Database as promiscuous. If a database is specified as promiscuous, the entries coming from this database will be treated differently during data unification. The entries coming from promiscuous databases, when unified, can belong to multiple User Entity s if they satisfy the equivalence conditions (imposed by the unification protocol) with any non-promiscuous entry belonging to the same set. A useful example of a promiscuous database is SCOP, database of protein structural domains, where a domain can be contained more than one protein (User Entity in our analogy).
See Figure 1.3 for a demonstration of how different unification approaches work in BIANA .
2. Installation
2.1 Requirements
- Windows:
- Cytoscape 2.6 (http://www.cytoscape.org/) (only required if BIANA is going to be used as a Cytoscape Plugin)
- MAC and Unix based systems:
- A MySQL server version 5+ (http://www.mysql.com/).
- Mac Users: If you experience problems installing MySQL Server, check the following link: http://www.brainfault.com/2008/04/18/install-python-mysql-mysqldb-on-mac-osx/.
- Python, version 2.5 http://www.python.org/
- Cytoscape 2.6 (http://www.cytoscape.org/) (required if BIANA is going to be used as a Cytoscape Plugin)
- A MySQL server version 5+ (http://www.mysql.com/).
2.2 Installation
- Windows:
- Download the BIANA Windows Installer and follow setup instructions.
- Installation of BIANA Cytoscape Plugin:
- Use the Plugin Cytoscape manager:
- Plugins
 Manage Plugins
Manage Plugins
- Change Download Site
 Edit sites
Edit sites
- Add http://sbi.imim.es/data/biana/Biana_cytoscape_plugin.xml).
- Plugins
- Execute Cytoscape and run BIANA Cytoscape plugin (Plugins
 BIANA).
BIANA).
- Automatically, it will ask you to select your Python Interpreter: Select the file biana.bat (located where you installed BIANA). If you want to change it in the future, go to Configuration
 Preferences.
Preferences.
- Use the Plugin Cytoscape manager:
- MAC and Unix based systems:
- Download source code from http://sbi.imim.es/web/BIANA.php.
- UNIX and MAC. Installation from Source Package:
- Installation WITH system administration privileges (which will install biana in site-packages of default Python interpreter):
{bianacommand} \$> ./install.sh - Installation WITHOUT system administration privileges:
- Use install.sh with destination path as an argument as follows:
{bianacommand} \$> ./install.sh <path\_to\_install\_biana> - Then update PYTHONPATH environment variable as follows (put it in .bash_profile to make this change permanent)
{bianacommand} \$> export PYTHONPATH=\$PYTHONPATH:<path\_to\_install\_biana>
- Use install.sh with destination path as an argument as follows:
- Installation WITH system administration privileges (which will install biana in site-packages of default Python interpreter):
- Checking whether installation was successful
To start using BIANA import biana library inside a Python script as follows:
{bianacommand} \$> python >>> import biana BIANA>
- Installation of BIANA Cytoscape Plugin:
- Use the Plugin Cytoscape manager:
- Plugins
 Manage Plugins
Manage Plugins
- Change Download Site
 Edit sites
Edit sites
- Add http://sbi.imim.es/data/biana/Biana_cytoscape_plugin.xml).
- Plugins
- Execute Cytoscape and run BIANA Cytoscape plugin (Plugins
 BIANA).
BIANA).
- Automatically, it will ask you to select your Python Interpreter: Select the python executable file in your system. If you want to change it in the future, go to Configuration
 Preferences.
Preferences.
- Use the Plugin Cytoscape manager:
3. Usage
BIANA is developed as a Python package, and it is thought to be executed in Python scripts. In order to facilitate users work, most of its commands can also be executed as a Cytoscape plugin.
3.1 Execution
3.1.1 Executing BIANA in a Python script
If you want to access to the BIANA Python module directly, you have to import it in your Python script:
{bianacommand}
import biana
or
{bianacommand}
from biana import *
For more details in BIANA available functionalities and methods, see sections 4 and 5. Some of the most common scripts are demonstrated in the scripts directory.
3.2 Executing BIANA as a Cytoscape plugin
On the Cytoscape menus at the top, go to Plugins and then click on BIANA.
{createbianadatabase}
create_biana_database( dbname = "YOUR_BIANA_DATABASE_NAME",
dbhost = "MYSQ_SERVER_HOST",
dbuser = "USER",
dbpassword = "PASSWORD",
description = "Test database" )
{createbianadatabasecommandline}
scripts/administration> python create_new_biana_database.py
{createbianadatabaseparameters}
*dbname: name of the new \Bdb. It must not exist previously!
*dbhost: name or IP where the MySQL server is.
*dbuser: user of MySQL database. User must have database creation grants!
*dbpassword: Password of MySQL user.
*description: Description for \Bdb
{createunification}
create_unification_protocol( dbname = "BIANA_DATABASE_V1",
dbhost = "localhost",
dbuser = "root",
dbpassword = "",
unification_protocol_name = "NAME",
list_unification_atom_elements = [([dbID_list],[attributes]),
([dbID_list],[attribute_list])] )
{createunificationcommandline}
scripts/administration> python create_unification_protocol.py
{createunificationparameters}
*unification_protocol_name: Name defining the unification protocol
*list_unification_atom_elements: List of tuples.
First position in each tuple consists of a list of external database identifiers, and
second position in the tuple consists in a list of external entity attributes to be used.
{deleteunification}
delete_unification_protocol( dbname = "BIANA_DATABASE",
dbhost = "MYSQL SERVER HOST",
dbuser = "USER",
dbpassword = "PASSWORD",
unification_protocol_name = ``UNIFICATION NAME'')
{deleteunificationcommandline}
scripts/administration> python delete_unification_protocol.py
{deletebianadatabase}
delete_biana_database( dbname = "DATABASE NAME",
dbhost = "MYSQL_SERVER_HOST",
dbuser = "USER",
dbpassword = "PASSWORD" )
{deletebianadatabasecommandline}
scripts/administration> python delete_biana_database.py
{parserscommandline}
scripts/administration> python parse_database.py
{startsession}
session = create_new_session( sessionID = "ID",
dbname="BIANA_DBNAME",
dbhost="BIANA_DBHOST",
dbuser="BIANA_DBUSER",
dbpassword="BIANA_DBPASS",
unification_protocol="No unification" )
{startsessionparameters}
*unification_protocol: Name defining the unification protocol to be used
{duplicateuserentityset}
session.duplicate_user_entity_set( user_entity_set_id="User_Entity_id1",
new_user_entity_set_id="New id" )
{removeuserentityset}
session.remove_user_entity_set( user_entity_set_id="User_Entity_Set_1" )
{selectalluserentities}
session.select_all_user_entities( user_entity_set_id = "User_Entity_Set_Name" )
{selectbylevel}
user_entity_set = session.get_user_entity_set(
user_entity_set_id = User_Entity_Set_Name")
user_entity_ids = user_entity_set.get_user_entity_ids(level = 1)
session.select_user_entities_from_user_entity_set(
user_entity_set_id = "User_Entity_Set_Name",
user_entity_id_list = user_entity_ids,
clear_previous_selection = True )
{selectbyattribute}
session.select_user_entities_from_user_entity_set(
user_entity_set_id = "User_Entity_Set_Name",
identifier_description_list =
[ ("attr_name", "attr_value"), ... ],
external_entity_attribute_restriction_list =
[ ("attr_name", "attr_value"), ... ],
id_type = "embedded",
clear_previous_selection = True )
{unselectuserentities}
user_entity_set = session.get_user_entity_set(
user_entity_set_id = "User_Entity_Set_Name")
user_entity_set.clear_user_entity_selection()
{taguserentities}
session.tag_selected_user_entities( user_entity_set_id = "User_Entity_Set_Name",
tag = "tag_name" )
{deleteuserentities}
user_entity_set = session.get_user_entity_set(
user_entity_set_id = "User_Entity_Set_Name")
user_entity_set.select_user_entities(
user_entity_id_list = [ (id_user_entity_1,
id_user_entity_2, ... ] )
session.remove_selected_user_entities(
user_entity_set_id = "User_Entity_Set_Name" )
{subuserentityset}
user_entity_set = session.get_user_entity_set(
user_entity_set_id = "User_Entity_Set_Name")
user_entity_set.select_user_entities(
user_entity_id_list = [ (id_user_entity_1,
id_user_entity_2, ... ] )
session.get_sub_user_entity_set(
user_entity_set_id = "User_Entity_Set_Name",
include_relations = True,
new_user_entity_set_id = "New_User_Entity_Name" )
{intersection}
session.get_intersection_of_user_entity_set_list(
user_entity_set_list = [ "User_Entity_Set_Name_1",
"User_Entity_Set_Name_2", ... ]
include_relations = True,
new_user_entity_set_id = "New_User_Entity_Name" )
{union}
session.get_union_of_user_entity_set_list(
user_entity_set_list = [ "User_Entity_Set_Name_1",
"User_Entity_Set_Name_2", ... ]
include_relations = True,
new_user_entity_set_id = "New_User_Entity_Name" )
{outputuserentities}
session.output_user_entity_set_details (
user_entity_set_id = "User_Entity_Set_Name",
attributes = [ "attr_name_1", "attr_name_2", ... ],
only_selected = False,
output_format = "xml",
out_method = sys.stdout.write() )
{outputexternalentities}
session.output_external_entity_details (
user_entity_id_list = [ "external_etity_1",
"external_entity_2", ... ],
attributes = [ "attr_name_1", "attr_name_2", ... ],
outmethod = sys.stdout.write() )
{createnetwork}
session.create_network(user_entity_set_id,
level=0,
include_relations_last_level = True,
relation_type_list=[],
relation_attribute_restriction_list=[],
use_self_relations=True,
expansion_attribute_list=[],
expansion_relation_type_list=[],
expansion_level=2,
attribute_network_attribute_list=[],
group_relation_type_list=[])
{createnetworkparameters}
* user_entity_set_id: identifier of user entity set for
which network will be created
* level: level of the network to be created,
network will be expanded till that level
* include_relations_last_level: include relations between the nodes
residing at the last level
* relation_type_list: type of the relations to be used in expansion
* relation_attribute_restriction_list: tuples of (attribute, value)
corresponding to restrictions to be applied on attributes of relations
* use_self_relations: include relations within the node itself
* expansion_attribute_list: tuples of (attribute, value_dictionary) corresponding
to attributes to be used in relation inference between nodes based on
shared attributes - value_dictionary is empty if attribute is not parameterizable
* expansion_relation_type_list: type of relations to be used in shared attribute
based relation inference
* expansion_level: number of relations (edges) to
look further while inferring relations based on shared attributes
* attribute_network_attribute_list: tuples of (attribute, value) corresponding to
attributes to be used while associating nodes with common attributes - value_dictionary
is empty if attribute is not parameterizable
* group_relation_type_list: type of relations that are going to be treated
as a group (like pathway, complex, cluster..)
{randomizenetwork}
session.create_randomized_user_entity_set(user_entity_set_id,
new_user_entity_set_id,
type_randomization)
{randomizenetworkparameters}
* user_entity_set_id: id of the user entity set whose
copy with random network is going to be created
* new_user_entity_set_id: id for the created
copy of user entity set
* type_randomization: randomization type to be used in network randomization,
can be one of the following: "random", "preserve_topology", "preserve_topology_and_node_degree",
"preserve_degree_distribution", "preserve_degree_distribution_and_node_degree", "erdos_renyi", "barabasi_albert"
where;
- "random": add same number of edges randomly between nodes of original graph
- "preserve_topology": keep edges, shuffle nodes of original graph
- "preserve_topology_and_node_degree": keep edges, shuffle nodes of original graph with the nodes of same degree
- "preserve_degree_distribution": remove an edge between two random nodes with degrees k, l then add to two nodes with degrees k-1 & l-1, then shuffle nodes
- "preserve_degree_distribution_and_node_degree": remove 2 random edges between a-b and c-d where degree(a)=degree(c) and degree(b)=degree(d) then add 2 edges between a-d and b-c, then shuffle nodes with the same degree
- "erdos_renyi": creates a graph where edges are redistributed based on erdos renyi random model
- "barabasi_albert": creates a graph where edges are redistributed based on barabasi albert model (preferential attachment)
{outputnetworkdetails}
session.output_user_entity_set_network(user_entity_set_id,
out_method=None,
node_attributes = [],
participant_attributes = [],
relation_attributes=[],
allowed_relation_types="all",
include_participant_tags=True,
include_relation_tags=True,
include_relation_ids=True,
include_participant_ids=True,
include_relation_type=True,
include_relation_sources=True,
output_1_value_per_attribute=True,
output_format="xml",
value_seperator=", ",
only_selected=False,
include_command_in_rows=False,
substitute_node_attribute_if_not_exists=False,
include_unconnected_nodes=True)
{outputnetworkdetailsparameters}
* output_1_value_per_attribute: Boolean. Defines whether 1 or multiple values
are outputted per each attribute
* output_format: format for the output used in case format is "table"; can be "tabulated" or "xml"
* include_relation_ids: Boolean to whether display or not relation identifiers
* include_participant_ids: Boolean to whether display or not relation participant identifiers
* include_relation_type: Boolean to whether display or not types of relations
* include_relation_sources: Boolean to whether display or not relation sources
* include_participant_tags: Boolean to whether display or not tags of participants
* include_relation_tags: Boolean to whether display or not tags of relations
* value_separator: string to separate consecutive values in the same column
* only_selected: Boolean to decide whether to output only selected nodes or all nodes (and their interactions)
* include_command_in_rows: Include the command to output individual relation information at each row
* substitute_node_attribute_if_not_exists: In case the node does not have a value for a
given attribute (s.t. uniprotaccession) this flag make it possible to output another
attribute (e.g. geneid) in the same column indicated as attribute:value (e.g. geneid:123123)
* include_unconnected_nodes: Boolean to whether display or not unconnected nodes
{outputrelationdetails}
session.output_external_entity_relation_details(out_method=None,
external_entity_relation_id_list=[],
attributes=[],
node_attributes=[],
relation_attributes=[],
participant_attributes=[])
{outputrelationdetailsparameters}
* external_entity_relation_id_list: list of relation identifiers
for which details will be outputted
* node_attributes: attributes of user entities connected by
these relations for which information will be fetched
* relation_attributes: attributes of external entity relations for
which information will be fetched
* participant_attributes: attributes of external entity relation participants for
which information will be fetched
* out_method: output method to be used if None overwritten by instance default output method
4. BIANA administration commands
BIANA administration commands consist of a set of procedures to create and maintain BIANA databases in a transparent way designated for end users. The user executing these administration commands must be granted MySQL permissions for creating new databases, new tables and inserting new data, updating, altering, deleting existing data, lock and drop tables depending on the command.
BIANA uses a MySQL database in order to store information obtained from external databases. BIANA offers to the user a transparent way of creating and managing distinct BIANA Databases, in which each one can contain distinct external databases and distinct unification protocols.
The basic procedure when installing BIANA , is the following:
- Install BIANA and all its requirements.
- Create a new BIANA Database.
- Populate BIANA Database by inserting information from external databases.
- Integrate external databases using a unification protocol decided by the user.
- Start working with the created database and unification protocol.
* Considerations to take in account:
In order to optimize data insertion and data access, BIANA Databasescan be found in two possible states:
- Parsing state: State in which database is optimized for parsing step. This is the default state when a new BIANA Databaseis created.
- Running state: State in which database is optimized for running step. Database changes to this state in the first running procedure.
4.1 Executing administration commands
4.1.1 From graphical interface
All administration options are found on BIANA Menu Configuration button.
4.1.2 From command line
All scripts related with administration commands are found on path:
{bianacommand}
biana/scripts/administration/script_to_execute {Tentative path}
4.1.3 From Biana API
All administration methods can be executed by using the Biana API:
{bianacommand}
import biana
biana.scripts.administration.COMMAND_TO_EXECUTE
4.2 Create a new BIANA database
- Function: Creates an empty BIANA repository and prepares this repository for parsing external databases. It creates all necessary tables and database indices.
- Special requirements: User must have database CREATION, LOCK and INSERT grants on MySQL server.
- Executing from Graphical Interface: Configuration
 Create new BIANA database
Create new BIANA database
- Executing it from BIANA API:
- Executing it from command line:
- Parameters:
4.3 Populate an existing BIANA Database
- Function: Inserts the information from an external database into selected BIANA Databaseusing selected parser.
- Special requirements: User must have INSERT grant on MySQL server for the selected BIANA Database.
- Executing from Graphical Interface:
Configuration
 Parse External Database
Parse External Database
- Executing it from BIANA API: Not possible. Use command line or graphical interface.
- Executing it from command line:
To check which external databases are available, which files are required and how to execute parsers using command line, see section 6.
4.4 Drop an existing BIANA Database
- Function: Drops an existing BIANA Database. Precaution! This action can not be undone! If you want to drop a BIANA Databasebut have a permanent copy in a smaller file, see section 7.1.
- Special requirements: User must have database DROP grants on MySQL server.
- Executing from Graphical Interface: Configuration
 Delete BIANA Databases
Delete BIANA Databases
- Executing it from BIANA API:
- Executing it from command line:
4.5 Create a new unification protocol in a BIANA Database
- Function: Creates a new unification protocol from scratch using a given BIANA Database.
- Special requirements: User must have INSERT, CREATE, DELETE and DROP grants on MySQL for selected BIANA Database.
- Executing from Graphical Interface: Configuration
 Create New Unification Protocol
Create New Unification Protocol
- Executing it from BIANA API:
- Executing it from command line:
- Parameters:
4.6 Drop an unification protocol in a BIANA Database
- Function: Drops an existing unification protocol in a BIANA Database. Precaution! This action can not be undone! If you want to drop an unification protocol in a BIANA Databaseit is recommended to have a permanent copy of the complete database in a file, see section ``Dump BIANA Database''.
- Special requirements: User must have DROP database grant on MySQL server.
- Executing from Graphical Interface: Configuration
 Delete Unification Protocol
Delete Unification Protocol
- Executing it from BIANA API:
- Executing it from command line:
5. BIANA working commands
5.1 Preliminary introduction
5.1.1 User Entity Concept Focused
BIANA works with the abstract concept of ``User Entity'', as explained in the introduction section. User entities are the collections of external entities that are considered to be equivalent according to a unification protocol. So, it is not possible to directly compare user entities coming from distinct unification protocols.
5.1.2 User Entity Set creation
The first step to create a network is the acquisition of an initial set of seed user entities (represented as nodes) (i.e. the biologic entities of interest). Note that if no unification is used, these user entities would correspond to individual biomolecules (like proteins and genes) other than a set of biomolecules. The created set of user entities is called User Entity Set. Once created, user can interact directly with one or more user entity sets by combining them via union or intersection. User entity sets are created from a given list of attribute and value pairs which describe biological entries in some data repository (e.g. (UniProt accession, P49137), (gene symbol, MAPKAPK2), (uniprot entry name, MAPK2_HUMAN), (HGNC, 6887), (HPRD, 11882), (Entrez, 9261), ...). All user entry possessing external entries associated with these values for specified attributes will be contained in the same user entity set. Some restrictions can also be imposed (for example, user may dictate that all user entities in the set must have the attribute Taxonomy Name ``human'').
5.1.3 Network expansion progressively by levels
Once the user has created the user entity set, it can be expanded to further levels, creating a network of relations. Relations can be of different types:
- User entity relations
- . Two user entities will be linked if any of their external entities have a relation observed in any external database. This includes interactions, reactions, pathways, ...). BIANA works only with the concept of BINARY relations, which are represented as edges. So, if user selects to view ``pathway'' relations, two nodes belonging to the same pathway will be linked by a direct edge. So, there will be an edge between all nodes belonging to the same pathway. This is applied to all types of relations (see group expansion to expand by some relations types without adding edges).
- Attribute relations
- . Two user entities will be linked if they share some attributes (for example, user can create a network of protein similarity, where nodes will be linked if they share sequence similarity measures).
- Predicted relations by attribute expansion
- . BIANA predicts novel relationships based on information transferred by using common properties shared by the nodes of the graph. Basically, let x, y, z be biological entities obtained with the integration approach, an interaction is predicted between x and y, if x is observed to interact with z and y shares some attributes (decided by the user, i.e. PFAM domains, sequence similarity, etc.) with node z. BIANA can help to unravel latent relationships between entities using various attributes such as sequence similarity using cutoffs based on e-value or percentage of identity, PFAM or SCOP domains, or GO terms.
5.2 Start a working session
- Function: Starts a new working session, using a specified Biana Database and given Unification Protocol.
- Special requirements: A populated Biana database must exist with the parameters given. User must have SELECT grant on MySQL server for the selected BIANA Database.
- Executing from Graphical Interface: New Session Button
- Executing it from BIANA API:
- Parameters:
The method returns a session object. The session object is also stored in a session dictionary, named available_sessions, where sessions are identified by their unique sessionIDs. So, session object can be accessed directly (session.METHOD_TO_EXECUTE) or by using the session dictionary (available_sessions["SessionID"].METHOD_TO_EXECUTE). In the following examples, the second way is used.
5.3 User Entity Sets: Characteristics and methods
The User Entities is the basic BIANA set of data to work. First it is necessary to create a Set by defining which entities must be in it, by selecting which attribute values and restrictions they must have. After creating the initial set, it can be extended to further levels by adding the relation partners of seed entities. So, a User Entity Set contains the seed user entities obtained with the attribute values given during set creation, and all the user entities obtained during network creation.
5.3.1 Create a new set of user entities
- Function: Creates a new set of user entities based on given user entity attribute values and user entity attribute restrictions.
- Special requirements: A working session has been started
- Executing from Graphical Interface: BIANA Session
 Create New Set.
Create New Set.
- Executing it from BIANA API:
- Parameters:
A User Entity Set is characterized by:
- The user entities it contains (nodes)
- The levels of their nodes (at which step of network extension nodes have been added)
- Relations between user entities (edges)
- Tags assigned to nodes and relations
- Groups of nodes by some criteria
In the graphical interface, these elements can be accessed by using the User Entity Set Tree, which has three elements:
- Network:
- All nodes contained in the user entity set classified by the level of relation.
- Tags:
- All tags added to nodes or edges.
- Groups:
Actions that can be performed in a user entity set can be accessed in the graphical interface by selecting the user entity set in the Session Tree. Using Biana API, all the methods on user entity sets must be used through the Session instance, where the user entity must be called as the user_entity_set_id parameter.
5.3.2 Duplicate a user entity set
- Function: Creates a new user entity set with another name by duplicating an existing one. It is an exact copy of the original one (same set restrictions, same levels, ...)
- Special requirements: A user entity set has been created.
- Executing from Graphical Interface: User Entity Set Tree Node
 Duplicate
Duplicate
- Executing it from BIANA API:
5.3.3 Remove a user entity set
- Function: Deletes a user entity set. This action is not reversible!
- Special requirements: The use entity set must previously exist.
- Executing from Graphical Interface: User Entity Set Tree Node
 Delete
Delete
- Executing it from BIANA API:
5.3.4 Select nodes in a user entity set
In most experiments, it is interesting to select a subset of nodes inside a User Entity Set because of several reasons: create a new user entity set with selected nodes, analyze a subset of nodes instead of all the nodes, etc. Nodes can be selected using distinct criteria:
- Nodes that have some attribute.
- Nodes belonging to certain level on the network.
- Mapping one user entity set on other (select the nodes that are in the intersection in both user entity sets).
- Manually selecting nodes in network viewer inside Cytoscape Plugin
- Directly by using User Entity ID.
- Function:
- Special requirements:
- Executing from Graphical Interface:
User Entity Set Tree Node Network
Network  Level X
User Entity Set Tree Node
Level X
User Entity Set Tree Node  Select User Entities by
User Entity Set Tree Node
Select User Entities by
User Entity Set Tree Node  Select All User Entities
Manually selecting nodes in network viewer inside Cytoscape Plugin
Select All User Entities
Manually selecting nodes in network viewer inside Cytoscape Plugin
- Executing it from BIANA API:
- To select all nodes of a user entity set:
- To select nodes by level:
- To select nodes by attribute:
- To select all nodes of a user entity set:
5.3.5 Clear previous selection of nodes in a user entity set
Care must be taken during selection actions, not clearing previous selections may cause undesired nodes included in the following actions. To clean previous selection of nodes:
- Function: Clears the selection of nodes in a user entity set.
- Special requirements: A user entity set has been created and has selected nodes.
- Executing from Graphical Interface: Simply click on the network where there are no nodes and edges, and the current selection will disappear.
- Executing it from BIANA API:
5.3.6 Tag selected nodes in a user entity set
Tags are used to mark a set of selected nodes in a given moment. For example, if a User Entity Set contains nodes related to some illnesses, it would be interesting to tag them to analyze their properties compared with the rest of the nodes. Tags are always applied to selected nodes.
- Function: Tags all selected nodes in a User Entity Set.
- Special requirements: A user entity set has been created and there are some user entities selected.
- Executing from Graphical Interface: Right-click one of the selected nodes in network viewer inside Cytoscape Plugin.
From the menu that appears, select;
BIANA
 Tag Selected Nodes.
Then in the pop-up dialog box, enter the name of the tag as shown below.
Tag Selected Nodes.
Then in the pop-up dialog box, enter the name of the tag as shown below.
- Executing it from BIANA API:
5.3.7 Delete selected nodes in a user entity set
- Function: A selected group of nodes can be removed from the network they belong to permanently. Deletes selected user entities and their relations in the user entity set.
- Special requirements: A user entity set is created and it has selected nodes.
- Executing from Graphical Interface: Right-click one of the selected nodes in network viewer inside Cytoscape Plugin.
From the arising menu, select;
BIANA
 Remove Selected Nodes. Then in the pop-up dialog box, select ``Yes'', to accept the removal irreversibly as demonstrated below.
Remove Selected Nodes. Then in the pop-up dialog box, select ``Yes'', to accept the removal irreversibly as demonstrated below.
- Executing it from BIANA API:
5.3.8 Create a sub user entity set
More often than not, users may be interested in generating a new user entity set from a subset of nodes in an existing user entity set.
- Function: Creates a new set from selected user entities in a user entity set. The new set DOES not include any of the attribute restrictions from its parent set.
- Special requirements: A user entity set is created and it has selected user entities.
- Executing from Graphical Interface: Right click one of the selected nodes in network viewer inside Cytoscape Plugin.
From the arising menu, select;
BIANA
 Create new set from selected nodes.
Specify the name for the user entity set to be created from the appearing dialog box;
Create new set from selected nodes.
Specify the name for the user entity set to be created from the appearing dialog box;
- Executing it from BIANA API:
5.3.9 Intersection of user entity sets
Any number of user entity sets can be intersected with each other to generate a new user entity set containing nodes and edges common to all of them.
- Function: Gets a new user entity set containing the user entities that are in the intersection of all user entity sets specified.
- Special requirements:
- Executing from Graphical Interface:
- Select more than one user entity set from BIANA Session Tree (click while holding control key) and right-click.
- From the arising menu, select ``Intersection''; BIANA Session Tree Selected User Entity Nodes
 Intersection
Intersection
- Specify the name for the user entity set to be created from the appearing dialog box;
- Executing it from BIANA API:
5.3.10 Union of user entity sets
Any number of user entity sets can be combined with each other to generate a new user entity set containing nodes and edges in all of them.
- Function:
- Special requirements:
- Executing from Graphical Interface:
- Select more than one user entity set from BIANA Session Tree (click while holding control key) and right-click.
- From the arising menu, select ``Union''; BIANA Session Tree Selected Nodes
 Union
Union
- Specify the name for the user entity set to be created from the appearing dialog box;
- Executing it from BIANA API:
5.3.11 View and Export
5.3.11.1 Output user entity details from a user entity set
- Function:Shows or prints into a file user entities information.
- Executing from Graphical Interface:
- All user entities in the set:
- BIANA Session Tree Selected User Entity Node
 View Set Details
View Set Details
- Select attributes for which information of user entities will be retrieved from the arising window.
- BIANA Session Tree Selected User Entity Node
- Only selected user entities:
- Select a set of nodes and right-click one of them.
- In the appearing menu: BIANA
 View Entity Details-
View Entity Details-
- Select relevant attributes from the pop-up window as explained in the previous option.
- All user entities in the set:
- Executing it from BIANA API:
5.3.11.2 Show user entity attributes
Prints the composition details from selected User Entities, where attributes are showed for each External Entity .
- Function: Shows or prints into a file External Entities information that belong to selected User Entities.
- Executing from Graphical Interface:In the user entity set details window, it is possible to select and display information about external entities contained in individual user entities by;
User Entity Set Details Window
 View Details
View Details
- Executing it from BIANA API:
5.3.12 Creating a network in a User Entity Set
BIANA allows user to create 3 different types of relation networks:- Relation network:
- connecting user entities (nodes) with respect to individual relationships (interaction, pathway, reaction, no_interaction) of contained external entities of user entities.
- Attribute network:
- connecting user entities with respect to shared attributes of contained external entities of user entities.
- Expansion network:
- connecting user entities with respect to predicted relations based on some attributes.
- Function: Creates a network of relations
- Special requirements: A user entity set has been created.
- Executing from Graphical Interface: User Entity Set Tree Node
 Create/Expand Network or User Entity Set Tree Node Network
Create/Expand Network or User Entity Set Tree Node Network  Create/Expand. Then in the network selection window (Image 20), check ``Add attribute relations''. Next, in the appearing pop-up window, select types of relations between user entities to be added and restrictions to be applied on those relations.
Create/Expand. Then in the network selection window (Image 20), check ``Add attribute relations''. Next, in the appearing pop-up window, select types of relations between user entities to be added and restrictions to be applied on those relations.
- Executing it from BIANA API:
- Parameters:
ATTENTION! The attribute value list in expansion_attribute_list argument is reserved to be used for attribute type ``proteinSequence'' and should be empty ( [] ) for attributes other than ``proteinSequence''. In case of ``proteinSequence'' attribute valid options are ``identities'', ``similarity'', ``coverage_A'', ``coverage_B'', ``bit_score'', ``evalue''.
5.3.13 Network Randomization
- Function: Randomizes current network in user entity set.
- Executing from Graphical Interface:User Entity Set Tree Node
 Randomize Network
or
User Entity Set Tree Node Network
Randomize Network
or
User Entity Set Tree Node Network  Randomize
Randomize
- Executing it from BIANA API:
- Parameters:
5.3.14 Output network of relations
- Function: Shows or prints into a file user entities network information.
- Executing from Graphical Interface: User Entity Set Tree Node
 View Network Details or User Entity Set Tree Node Network
View Network Details or User Entity Set Tree Node Network  View Details. Then in the attribute selection window, select attributes for which information of edges and nodes connected by those edges will be retrieved.
View Details. Then in the attribute selection window, select attributes for which information of edges and nodes connected by those edges will be retrieved.
- Executing it from BIANA API:
- Parameters:
5.3.15 Show relation details
- Function: Shows or prints into a file the details of a user entity relation.
- Executing from Graphical Interface: In the user entity set network details window, it is possible to select and display information about all relations connecting external entities contained in individual user entities by selecting relations from the user entity network details view window and clicking ``€śView Details''€ť button yielding in a new information window poped-up. User Entity Set Network Details Window
 View Details
View Details
- Executing it from BIANA API:
- Parameters:
6. BIANA External Databases Parsers
BIANA provides some default database parsers for most common databases and formats. BIANA has been designed to be able to store any kind of biologic database, relying on the user how he wants to integrate data between databases by choosing which combinations of attributes must be shared. However, due to the large number of different databases, formats and versions, and that often different versions of the same database have different formats, not all databases with biologic data have a current working BIANA Parser. Despite existing interchange standard formats, databases often change their formats, so parsers are not guaranteed to work in all database versions. In order to solve this problem, we provide a set of default parsers, that will be updated in this list of available parsers.
If you find an existing parser is not working any more for a new database version, or you are interested in having a parser for another database not available here, you can ask for us to make it (it can take some time), or try yourself creating a new parser (see 6.3. Alternatively, you can use BIANA Generic Parser which accepts data in a certain tab-separated format (see 6.2). Once you convert your data you can user the Generic Parser to parse your data.
6.1 Available External Databases Parsers
An external database is any data source that contains biologic or chemical data that can be parsed by BIANA and inserted in the database in order to be integrated with data in other databases. Here are described available external database parsers, with the following information:
- External database.
- External database description.
- Needed external database files and how obtaining them.
- External entity description and external entity attributes.
- Last checked version.
- Approximate parsing time.
Distinct versions or releases of external databases may contain distinct formats or special characteristics than make parser to not work properly. If any error is produced during parsing due to unexpected database format, a control process is executed and whole database is deleted from BIANA Database. However, it is recommended to create a testing BIANA Databaseto inserted data before inserting it to the desired BIANA Database, in order to check all is working properly. Parsers can be used directly from Graphical Interface or by command line. A script for parsing is available in BIANA administration scripts.
6.1.1 Data retrieval
Data from external databases can be obtained directly from website or FTP links listed below. In order to facilitate data retrieval, in administration scripts directory there are several ftp scripts to automatically get the data.
{get_edb_by_ftp}
scripts/administration/external_database_download_scripts
Each script has one of the following formats:
- ftp_DATABASE
- wget_DATABASE
- html_DATABASE
To execute them, execute the following command, replacing DATABASE for the desired database depending on the prefix (either ftp, wget or html) preceding the DATABASE.
- For DATABASEs preceded with ftp:
{get_edb_by_ftp} \$> ftp_get_database.sh ftp_DATABASE # calls: ftp -inp < ftp_DATABASE - For DATABASEs preceded with wget:
{get_edb_by_ftp} \$> wget_get_database.sh ftp_DATABASE # calls: wget -i wget_DATABASE - For DATABASEs preceded with html: Those DATABASEs either requires registration or needs human interaction during download so use a web browser to go to the page indicated in html_DATABASE file.
{get_edb_by_ftp} \$> html_get_database.sh html_DATABASE # calls: lynx html_DATABASE
6.1.2 Available Parsers
6.1.2.1 Uniprot
| Description | Swiss-Prot, which is manually annotated and reviewed. TrEMBL, which is automatically annotated and is not reviewed. UniProt (Universal Protein Resource) is the world's most comprehensive catalog of information on proteins. It is a central repository of protein sequence and function created by joining the information contained in Swiss-Prot, TrEMBL, and PIR. |
| Database Reference | The UniProt Consortium (2007) The Universal Protein Resource (UniProt). Nucleic Acids Res. 35: D193-197. |
| Database Link | ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete |
| External Entity Types | Protein |
| External Entity Relation Types | |
| Needed files | uniprot_sprot.dat.gz (SWISS-PROT) and uniprot_trembl.dat.gz (TREMBL) |
| Parser name | uniprot |
| Input-identifier | Files uniprot_sprot.dat.gz and uniprot_trebml.dat.gz |
| Checked version | Uniprot Knowledgbase Release 14.1 (September 2008) |
| Comments | Swiss-prot and TrembL must be inserted as distinct databases. |
| Approximate parsing time | Swissprot: One hour (or less). Trembl: 8 hours |
Shell Command:
{parser_command}
\$> python parse_database.py uniprot --input-identifier=uniprot_sprot.dat.gz
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="uniprot swissprot"
--database-version="Release XX"
\$> python parse_database.py uniprot --input-identifier=uniprot_trembl.dat.gz
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="uniprot trembl"
--database-version="Release XX"
6.1.2.2 GenBank Database
| Description | GenBank is the NIH genetic sequence database, an collection of all publicly available DNA. GenPept contains proteins codified by those sequences. |
| Database Reference | Genbank. Nucleic Acids Res. 2007 Jan;35(Database issue):D21-5. |
| Database Link | ftp://ftp.ncbi.nih.gov/ncbiasn1/protein_fasta/ |
| External Entity Types | Protein |
| External Entity Relation Types | - |
| Needed files | All fsa_aa.gz files in ftp site |
| Parser name | ncbi_genpept |
| Input-identifier | The path where all fsa_aa.gz files are saved |
| Checked version | Release 167 |
| Comments | It is necessary to have previously inserted taxonomy database. |
| Approximate parsing time | 5 hours |
Shell Command:
{parser_command}
\$> python parse_database.py ncbi_genpept --input-identifier=path/
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="genpept"
--database-version="Release XX"
6.1.2.3 Taxonomy Database
| Description | The NCBI taxonomy database contains the names of all organisms that are represented in the genetic databases with at least one nucleotide or protein sequence |
| Database Reference | Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, Schuler GD, Tatusova TA, Rapp BA (2000). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2000 Jan 1;28(1):10-4 |
| Database Link | ftp://ftp.ncbi.nih.gov/pub/taxonomy |
| External Entity Types | Taxonomy |
| External Entity Relation Types | |
| Needed files | taxdump.tar.Z |
| Parser name | taxonomy |
| Input-identifier | Path where taxdump.tar.Z is uncompressed |
| Checked version | August 2008 |
| Comments | Uncompress and untar taxdump.tar.Z file |
| Approximate parsing time | 10 minutes |
Shell Command:
{parser_command}
\$> python parse_database.py taxonomy --input-identifier=path/
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="Taxonomy"
--database-version="Database release"
6.1.2.4 Protein-protein interactions Open Biomedical Ontologies
| Description | A structured controlled vocabulary for the annotation of experiments concerned with protein-protein interactions. Developed by the HUPO Proteomics Standards Initiative. |
| Database Reference | |
| Database Link | http://psidev.sourceforge.net/mi/psi-mi.obo |
| External Entity Types | PsiMiOboOntologyElement |
| External Entity Relation Types | - |
| Needed files | psimiobo |
| Parser name | psi_mi_obo |
| Input-identifier | Path where psi-mi.obo file is |
| Checked version | July 2008 |
| Comments | |
| Approximate parsing time | Less than a minute |
Shell Command:
{parser_command}
\$> python parse_database.py psi_mi_obo --input-identifier=path where psi-mi.obo file is
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="PSI-MI Obo"
--database-version="Database release"
6.1.2.5 NCBI Blast Non-Redundant Database
| Description | NCBI Non-redundant sequence database for Blast |
| Database Reference | Genbank. Nucleic Acids Res. 2007 Jan;35(Database issue):D21-5 |
| Database Link | ftp://ftp.ncbi.nih.gov/ncbi-asn1/protein_fasta/ |
| External Entity Types | Protein |
| External Entity Relation Types | |
| Needed files | nr.gz |
| Parser name | nr |
| Input-identifier | nr.gz file |
| Checked version | August 2008 |
| Comments | It is necessary to have previously inserted taxonomy database |
| Approximate parsing time | 4-5 hours |
Shell Command:
{parser_command}
\$> python parse_database.py nr --input-identifier=nr.gz
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="nr"
--database-version="Database release X"
6.1.2.6 International Protein Index Database (IPI)
| Description | The International Protein Index: An integrated database for proteomics experiments. |
| Database Reference | TKersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. The International Protein Index: An integrated database for proteomics experiments. Proteomics 4(7): 1985-1988 (2004). |
| Database Link | ftp://ftp.ebi.ac.uk/pub/databases/IPI/current/ |
| External Entity Types | Protein |
| External Entity Relation Types | |
| Needed files | All fasta files in ftp site |
| Parser name | ipi |
| Input-identifier | The path where all ipi.XXXX.fasta.gz files are saved |
| Checked version | 1 September 2008 |
| Comments | |
| Approximate parsing time | 10 minutes |
Shell Command:
{parser_command}
\$> python parse_database.py ipi --input-identifier=path_where_ipi_files_are
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="ipi"
--database-version="Database release X"
6.1.2.7 iRefIndex
| Description | A reference index for protein interaction data |
| Database Reference | iRefIndex: a consolidated protein interaction database with provenance. Razick S, Magklaras G, Donaldson IM. BMC Bioinformatics. 2008 Sep 30;9:405. |
| Database Link | ftp://ftp.no.embnet.org/irefindex/data/ |
| External Entity Types | Protein |
| External Entity Relation Types | Interaction and complex |
| Needed files | File ALl.mitab.??????.txt.zip |
| Parser name | iRefIndex |
| Input-identifier | The downloaded file |
| Checked version | 06042009 |
| Comments | |
| Approximate parsing time | 10 minutes |
Shell Command:
{parser_command}
\$> python parse_database.py irefindex --input-identifier=path_where_downloaded_file_is
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="iRefIndex"
--database-version="Database release X"
6.1.2.8 Cluster Of Orthologous Genes Database (COGs)
| Description | Clusters of Orthologous Groups of proteins (COG). COG is delineated by comparing protein sequences encoded in complete genomes, representing major phylogenetic lineages. Each COG consists of individual proteins or groups of paralogs from at least 3 lineages and thus corresponds to an ancient conserved domain. |
| Database Reference | Science 1997 Oct 24;278(5338):631-7, BMC Bioinformatics 2003 Sep 11;4(1):41. |
| Database Link | ftp://ftp.ncbi.nih.gov/pub/COG/COG/ |
| External Entity Types | Protein |
| External Entity Relation Types | |
| Needed files | myva, myva=gb, org.txt, fun.txt |
| Parser name | cog |
| Input-identifier | Path where cog files are downloaded |
| Checked version | 2003 |
| Comments | |
| Approximate parsing time | 165 seconds |
Shell Command:
{parser_command}
\$> python parse_database.py cog --input-identifier=path_where_cog_files_are
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="COG"
--database-version="Database release X"
--promiscuous
6.1.2.9 HUGO Gene Nomenclature Committee (HGNC)
| Description | HUGO Gene Nomenclature Committee. Each symbol is unique and we ensure that each gene is only given one approved gene symbol. |
| Database Reference | |
| Database Link | http://www.genenames.org/data/gdlw_index.html |
| External Entity Types | Protein |
| External Entity Relation Types | |
| Needed files | ``All data'' in ``Text format'' |
| Parser name | hgnc |
| Input-identifier | The file where the data is saved |
| Checked version | September 2008 |
| Comments | |
| Approximate parsing time | Less than a minute |
Shell Command:
{parser_command}
\$> python parse_database.py hgnc --input-identifier=FILE_WHERE_DATA_IS_SAVED
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="HGNC"
--database-version="Database release X"
6.1.2.10 KEGG: Kyoto Encyclopedia of Genes and Genomes
| Description | KEGG: Kyoto Encyclopedia of Genes and Genomes |
| Database Reference | KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27-30 (2000) |
| Database Link | ftp://ftp.genome.jp/pub/kegg/ |
| External Entity Types | protein, drug, compound, enzyme, glycan |
| External Entity Relation Types | relation, pathway, cluster |
| Needed files | ko (for kegg_ko) genes.tar.gz (for kegg_gene) compound drug enzyme glycan reaction (for keg_ligand) |
| Parser name | kegg_ko kegg_gene kegg_ligand |
| Input-identifier | ko (for kegg_ko) genes.tar.gz (for kegg_gene) |
| Checked version | |
| Comments | |
| Approximate parsing time |
Shell Command for Kegg KO:
{parser_command}
\$> python parse_database.py kegg_ko --input-identifier=FILE_WHERE_DATA_IS_SAVED
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="KEGG KO"
--database-version="Database release X"
Shell Command for Kegg Gene:
{parser_command}
\$> python parse_database.py kegg_gene --input-identifier=genes.tar.gz
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="KeggGene"
--database-version="Database release X"
Instead you can insert gene data in two steps using genes.pep and genes.nuc Peptide sequences:
{parser_command}
\$> python parse_database.py kegg_gene --input-identifier=genes.pep
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="KeggGenePep"
--database-version="Database release X"
{parser_command}
\$> python parse_database.py kegg_gene --input-identifier=genes.nuc
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="KeggGeneNuc"
--database-version="Database release X"
6.1.2.11 PSI-MI 2.5 Formatted databases
| Description | Protein protein interaction databases in PSI-MI 2.5 Format |
| Database Reference | |
| Database Link | |
| External Entity Types | |
| External Entity Relation Types | |
| Needed files | |
| Parser name | psi_mi_2.5 |
| Input-identifier | |
| Checked version | |
| Comments | |
| Approximate parsing time |
Shell Command:
{parser_command}
\$> python parse_database.py psi_mi_2.5 --input-identifier=FILE_WHERE_DATA_IS_SAVED
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="Database name"
--database-version="Database release X"
This parser has been tested for PSI-MI 2.5 formatted databases:
- IntAct
- Website
- http://www.ebi.ac.uk/intact/site/index.jsf
- Download
- all xml files in ftp://ftp.ebi.ac.uk/pub/databases/intact/current/psi25/species
- Input identifier
- Path containing the xml files.
- Checked
- for version of September 2008
- Parsing time
- 1/2 hour
- BioGrid
- Website
- http://www.thebiogrid.org/downloads.php
- Download
- BIOGRID-ORGANISM-x.x.xx.psi25.zip. It is necessary to uncompress manually the zip file (
unzip BIOGRID-ORGANISM-x.x.xx.psi25.zip) - Input identifier
- Path containing uncompressed files (usually it uncompresses it into a folder named ``textfiles'').
- Checked
- for version 2.0.44 (September 2008)
- Parsing time
- 1000 seconds
- DIP
- Website
- http://dip.doe-mbi.ucla.edu/ (It is necessary to register in order to download data.)
- Download
- FULL file from downloads, which is complete DIP data set (file dip200xxxxxxx.mif25).
- Input identifier
- dip200xxxxxxx.mif25 file
- Checked
- for version 2008.07.08
- Parsing time
- 250 seconds
- HPRD
- Website
- www.hprd.org
- Download
- HPRD_PSIMI_xxxxxx.tar.gz. Untar and unzip the file
- Input identifier
- Path where all uncompressed files are
- Checked
- Release 7
- Parsing time
- 746 seconds
- MPACT
- Description
- Currently MPact gives access to yeast protein-protein interaction data contained in CYGD.
- Website
- http://mips.gsf.de/genre/proj/mpact/
- Download
- file ftp://ftpmips.gsf.de/yeast/PPI/mpact-complete.psi25.xml.gz
- Input identifier
- File
- Checked
- version April 2007
- Parsing time
- 150 seconds
- MINT
- Website
- http://mint.bio.uniroma2.it/mint/download.do
- Download
- ftp://mint.bio.uniroma2.it/pub/release/psi/2.5/2008-05-21/dataset/full.psi25.zip file. Uncompressed it
- Input identifier
- Path where file is uncompressed
- Checked
- for version: 2008.05.21
- Parsing time
- less than an hour
6.1.2.12 Biopax Level 2 Formatted databases
| Description | BioPAX Level 2 covers metabolic pathways, molecular interactions and protein post-translational modifications |
| Database Reference | |
| Database Link | |
| External Entity Types | protein |
| External Entity Relation Types | interaction, pathway |
| Needed files | |
| Parser name | biopax_level_2 |
| Input-identifier | |
| Checked version | |
| Comments | |
| Approximate parsing time |
Shell Command:
{parser_command}
\$> python parse_database.py biopax_level_2 --input-identifier=FILE_WHERE_DATA_IS_SAVED
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass=ČPASSWORD"
--time-control
--database-name="Database name"
--database-version="Database release X"
This parser has been tested for Biopax Level 2 formatted databases:
- Reactome
- Website
- http://reactome.org/download/index.html
- Download
- ``Events in the BioPAX Level 2 format'' file. Uncompress this file
- Input identifier
- Path where uncompressed files are
- Checked
- September 2008
- Parsing time
- 700 seconds
6.1.2.13 Structural Classification of Proteins (SCOP)
| Description | SCOP. Structural Classification of Proteins |
| Database Reference | Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995). SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. |
| Database Link | http://scop.mrc-lmb.cam.ac.uk/scop/ |
| External Entity Types | protein domain |
| External Entity Relation Types | |
| Needed files | All parsable SCOP files |
| Parser name | scop |
| Input-identifier | Path where files are found |
| Checked version | 1.73 |
| Comments | Database-version must be exactly the same as the SCOP version (i.e. if it is release 1.73, database-version must be ``1.73''. |
| Approximate parsing time | 1 minute |
Shell Command:
{parser_command}
\$> python parse_database.py scop --input-identifier=PATH_WHERE_FILES_ARE_SAVED
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD""
--time-control
--database-name="SCOP"
--database-version="1.73"
--promiscuous
6.1.2.14 Protein Families database (PFAM)
| Description | The Pfam database is a large collection of protein families, each represented by multiple sequence alignments and hidden Markov models (HMMs). |
| Database Reference | The Pfam protein families database, Nucleic Acids Research (2008) Database Issue 36:D281-D288 |
| Database Link | http://www.geneontology.org/GO.downloads.shtml |
| External Entity Types | pattern, protein |
| External Entity Relation Types | |
| Needed files | pfamA-file-name=Pfam-A.full.gz pfamB-file-name=Pfam-B.gz pfamSeq-file-name=pfamseq.gz |
| Parser name | pfam |
| Input-identifier | Path where files are found. |
| Checked version | 23.0 |
| Comments | Need to use the following additional arguments with the following values: pfamA-file-name=Pfam-A.full.gz pfamB-file-name=Pfam-B.gz pfamSeq-file-name=pfamseq.gz |
| Approximate parsing time |
Shell Command:
{parser_command}
\$> python parse_database.py pfam --input-identifier=PATH_WHERE_FILES_ARE_SAVED
-- pfamA-file-name=Pfam-A.full.gz
--pfamB-file-name=Pfam-B.gz
--pfamSeq-file-name=pfamseq.gz
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="PFAM"
--database-version="23.0"
--promiscuous
6.1.2.15 Gene Ontology (GO)
| Description | The Gene Ontology project provides a controlled vocabulary to describe gene and gene product attributes in any organism. |
| Database Reference | Gene Ontology: tool for the unification of biology. Nature Genet. (2000) 25: 25-29 |
| Database Link | http://www.geneontology.org/GO.downloads.shtml |
| External Entity Types | ontology |
| External Entity Relation Types | |
| Needed files | gene_ontology_edit.obo |
| Parser name | go_obo |
| Input-identifier | gene_ontology_edit.obo |
| Checked version | |
| Comments | This parser is for go_obo_v1.2 |
| Approximate parsing time |
Shell Command:
{parser_command}
\$> python parse_database.py go_obo --input-identifier="gene_ontology_edit.obo"
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="GO"
--database-version="VERSION X"
6.1.2.16 STRING
| Description | |
| Database Reference | |
| Database Link | http://string.embl.de/ |
| External Entity Types | protein |
| External Entity Relation Types | functional_association |
| Needed files | protein.aliases.v7.1.txt, protein.links.detailed.v7.1.txt.gz and protein.sequences.v7.1.fa.gz |
| Parser name | string |
| Input-identifier | Path where downloaded files are |
| Checked version | v7.1 |
| Comments | Database version must be the same as in the files! (i.e. v7.1) |
| Approximate parsing time |
Shell Command:
{parser_command}
\$> python parse_database.py go_obo --input-identifier="gene_ontology_edit.obo"
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="STRING"
--database-version="v7.1"
6.2 Preparing my data to use the generic parser
BIANA can parse any kind of user provided data given in tabulated (tab-separated) text format with its pre-specified Generic Parser.
| Description | User defined (generic) data parser |
| External Entity Types | Any |
| External Entity Relation Types | Any |
| Parser name | Any |
| Input-identifier | File path |
| Comments | See explanation below to see details about file format |
| Approximate parsing time | Depends on database size |
{parser_command}
\$> python parse_database.py generic
--input-identifier=PATH_WHERE_INPUT_FILE_RESIDES
--biana-dbname="BIANA_DB"
--biana-dbuser="root"
--biana-dbpass="PASSWORD"
--time-control
--database-name="MyClinicalExperiment08"
--database-version="1.0"
A typical input file for this parser lets the program aware which type of entry it is providing via use of two tags at the beginning of a line:
- @EXTERNAL_ENTITY_DATA
- @EXTERNAL_ENTITY_RELATION_DATA
Line with @EXTERNAL_ENTITY_DATA tag is for specifying that information for individual user data entries is going to be parsed, whereas line with @EXTERNAL_ENTITY_RELATION_DATA tells that user data relation entry information is going to be given thereafter. In the first occurrence of any of these tags in the input file, it is required that they are followed by a so called definition line describing the names of the columns of the data they are providing afterwards.
Definition line begins with several default columns followed by userdefined columns.For external entity data entries default columns in the definition line are ``id'' and ``type'' corresponding to internal identifier and type of the entry. On the other hand, for external entity relation data entries, default columns in the definition line are ``id'', ``interactor_id_list'' and ``type'' corresponding to identifiers of the two participants and the type of the relationship. Each column should be separated by at least one tab character (``![]() t'') and values inside columns should not include tab character. It is worth noting here that fields in relations can give information about individual participants rather than just relation using ``interactor_id: ATTRIBUTE. In case, a list of values need to be provided in the same column, values should be separated by ``
t'') and values inside columns should not include tab character. It is worth noting here that fields in relations can give information about individual participants rather than just relation using ``interactor_id: ATTRIBUTE. In case, a list of values need to be provided in the same column, values should be separated by ``![]() '' character. If a column has no value, this should be denoted by ``-'' character. The concept can be understood better on the input file format explanation given below.
'' character. If a column has no value, this should be denoted by ``-'' character. The concept can be understood better on the input file format explanation given below.
@EXTERNAL_ENTITY_DATA id type geneID chebI name 1 protein 1234 - protein A 2 protein 2314 - protein A2 3 protein 9999 - protein B 4 protein 1111 - protein C 5 protein 6778 - protein D 6 protein 1982 - protein E 7 protein 12178 - protein_X 8 gene 38111 - - 9 gene 2018 - - 100 protein 1001 - protein1001 101 protein 1002 - prot1002 102 protein 1003 - p1003 103 protein 1004 - p1004 104 protein 1005 - p1005 105 protein 1006 - p1006 106 protein 1007 - p1007 W compound - 15377 water B12 compound - 8843 Lactoflavin|Vitamin B2 C1 compound - 15422 Adenosine 5'-triphosphate C2 compound - 16761 Adenosine 5'-diphosphate C3 compound - 17621 Riboflavin-5-phosphate @EXTERNAL_ENTITY_RELATION_DATA id interactor_id_list type name method_ID participants:role R1 6|7 interaction - 18 6: bait| 7:prey R2 2|3 interaction - 18 - R3 3|4 interaction - 18 - R4 100|101|102|103 complex ABC complex 109 - R5 103|104 interaction - 109 - R6 104|105 interaction - 109 - R7 105|106 interaction - 109 - R8 4|105 interaction - 18 - R9 W|B12|C1|C2|C3 reaction reaction_sample - B12: substrate|C1: substrate|C2: substrate|C3: product|W: product R10 R4|R5|R6|R7 pathway first_pathway - - R11 R2|R3|R8 pathway pathway2 - - R12 R10|R11 pathway global pathway - -
6.3 Creating your own parser for your own data
All parsers written in BIANA inherits BianaParser class found in biana/BianaParser/bianaParser.py. To write your own parser you need to create a new Python class whose parent is BianaParser. Then all you need to define is the arguments your parser would require in the __init__ (class constructor) method and overwrite parse_database member method which is responsible from reading and inserting information from your data files.
Here is an example parser (MyDataParser.py) to insert data in user specified format into BIANA . Let's go over the code.
First we start with subclassing BianaParser:
{parser_command}
from bianaParser import *
class MyDataParser(BianaParser):
"""
MyData Parser Class
Parses data in the following format (meaining Uniprot_id1 interacts with Uniprot_id2 and
some scores are associated with both the participants and the interaction):
Uniprot_id1 Description1 Participant_score1 Uniprot_id2 Description2
Participant_score2 Interaction_Affinity_score
"""
name = "mydata"
description = "This file implements a program that fills up tables
in BIANA database from data in MyData format"
external_entity_definition = "An external entity represents a protein"
external_entity_relations = "An external relation represents an interaction with given affinity"
Above we introduce our parser and give name and description attributes, mandatory fields that are going to be used by BIANA to describe this parser. Then we create __init__ method where we call the constructor of the parent (BianaParser) with some additional descriptive arguments. You can add additional compulsory arguments to be requested from user by including "additional_compulsory_arguments" with a list of triplets (argument name, default value, description) (see list of command line arguments accepted by BianaParser by default).
{parser_command}
def __init__(self):
"""
Start with the default values
"""
BianaParser.__init__(self, default_db_description = "MyData parser",
default_script_name = "MyDataParser.py",
default_script_description = MyDataParser.description,
additional_compulsory_arguments = [])
Next, we are going to overwrite parse_database method (responsible from reading and inserting information from your data files) where we introduce some initial arrangements to let BIANA know about the characteristics of the data we are going to insert:
{parser_command}
def parse_database(self):
"""
Method that implements the specific operations of a MyData formatted file
"""
# Add affinity score as a valid external entity relation since it is not recognized by BIANA
self.biana_access.add_valid_external_entity_attribute_type( name = "AffinityScore",
data_type = "double",
category = "eE numeric attribute")
# Add score as a valid external entity relation participant attribute
# since it is not recognized by BIANA
# (Do not confuse with external entity/relation score attribute,
#participants can have their attributes as well)
self.biana_access.add_valid_external_entity_relation_participant_attribute_type(
name = "Score", data_type = "float unsigned" )
# Since we have added new attributes that are not in the default BIANA distribution,
#we execute the following command
self.biana_access.refresh_database_information()
There are various attributes and types in BIANA to annotate data entries coming from external databases (see attributes and types recognized by BIANA for details). In case we need to use attributes/types that are not by default recognized by BIANA we need to make them known to BIANA as it is done above with add_valid_external_entity_attribute_type and add_valid_external_entity_relation_participant_attribute_type methods (see defining new attributes and types for details).
{parser_command}
# Open input file for reading
self.input_file_fd = open(self.input_file, 'r')
# Keep track of data entries in the file and ids assigned by BIANA for them in a dictionary
self.external_entity_ids_dict = {}
for line in self.input_file_fd:
(id1, desc1, score1, id2, desc2, score2, score_int) = line.strip().split()
Above we open a file for reading and start reading the file. This is followed by converting data read from the file into objects BIANA will understand and insert them into database:
{parser_command}
# Create an external entity corresponding to Uniprot_id1 in database (if it is not already created)
if not self.external_entity_ids_dict.has_key(id1):
new_external_entity = ExternalEntity( source_database = self.database,
type = "protein" )
# Annotate it as Uniprot_id1
new_external_entity.add_attribute( ExternalEntityAttribute( attribute_identifier= "Uniprot",
value=id1, type="cross-reference") )
# Associate its description
new_external_entity.add_attribute( ExternalEntityAttribute( attribute_identifier= "Description",
value=desc1) )
# Insert this external entity into database
self.external_entity_ids_dict[id1] = \
self.biana_access.insert_new_external_entity( externalEntity = new_external_entity )
# Create an external entity corresponding to Uniprot_id2 in database (if it is not already created)
if not self.external_entity_ids_dict.has_key(id2):
new_external_entity = ExternalEntity( source_database = self.database, type = "protein" )
# Annotate it as Uniprot_id2
new_external_entity.add_attribute( ExternalEntityAttribute( attribute_identifier= "Uniprot",
value=id2, type="cross-reference") )
# Associate its description
new_external_entity.add_attribute( ExternalEntityAttribute( attribute_identifier= "Description",
value=desc2) )
# Insert this external entity into database
self.external_entity_ids_dict[id2] = self.biana_access.insert_new_external_entity( \
externalEntity = new_external_entity )
Finally we insert information of the interaction as follows:
{parser_command}
# Create an external entity relation corresponding to interaction between Uniprot_id1
# and Uniprot_id2 in database
new_external_entity_relation = ExternalEntityRelation( source_database = self.database,
relation_type = "interaction" )
# Associate Uniprot_id1 as the first participant in this interaction
new_external_entity_relation.add_participant( externalEntityID = \
self.external_entity_ids_dict[id1] )
# Associate Uniprot_id2 as the second participant in this interaction
new_external_entity_relation.add_participant( externalEntityID = \
self.external_entity_ids_dict[values[1]] )
# Associate score of first participant Uniprot_id1 with this interaction
new_external_entity_relation.add_participant_attributes( externalEntityID = \
self.external_entity_ids_dict[id1],
participantAttribute = ExternalEntityRelationParticipantAttribute( \
attribute_identifier = "Score",
value = score1 ) )
# Associate score of second participant Uniprot_id2 with this interaction
new_external_entity_relation.add_participant_attributes( externalEntityID = \
self.external_entity_ids_dict[id2],
participantAttribute = ExternalEntityRelationParticipantAttribute( \
attribute_identifier = "Score", value = score2 ) )
# Associate the score of the interaction with this interaction
new_external_entity_relation.add_attribute( ExternalEntityRelationAttribute( attribute_identifier = "AffinityScore",
value = score_int ) )
# Insert this external entity relation into database
self.biana_access.insert_new_external_entity( externalEntity = new_external_entity_relation )
As a good programming practice we do not forget to close the file we red as follows:
{parser_command}
self.input_file_fd.close()
6.4 Command line arguments accepted by parsers
By default BIANA parsers require:
- input-identifier=
- : path or file name of input file(s) containing database data. Path names must end with /.
- biana-dbname=
- : name of database biana to be used
- biana-dbhost=
- : name of host where database biana to be used is going to be placed
- database-name=
- : internal identifier name to this database (it must be unique in the database)
- database-version=
- : version of the database to be inserted"
The following optional arguments are also recognized:
- biana-dbuser=
- : user name for the specified host
- biana-dbpass=
- : password for the specified user name and host
- optimize-for-parsing
- : set to disable indices (if there is any) and reduce parsing time. Useful when you want to insert a considerable amount of data to an existing BIANA Databasewith indices created
- promiscuous
- : set to allow entries coming from parsed database to belong multiple User Entities.
6.5 Attributes and types recognized by BIANA and defining new ones
BIANA uses a set of attributes and types to define external entities coming from external biological databases (such as Uniprot Accession, STRING id, GO id, etc... as attributes and protein, DNA, interaction, complex, etc... as types). If you write a parser specialized for a particular data you have, you could either use existing attributes and types to annotate the entries in your data or create new ones if existing ones do not work for you. Here we give a list of valid BIANA attributes:
- External Entity & External Entity Relation Attributes
CHEBI COG CYGD DIP EC Encode Ensembl FlyBase GDB GeneID GeneSymbol GenomeReviews GI GO HGNC HPRD Huge IMGT IntAct IntEnz InterPro IPI KeggCode KeggGene Method_id MGI MIM MINT MIPS OrderedLocusName ORFName PFAM PIR PRINTS PRODOM Prosite psimi_name PubChemCompound Ratmap Reactome RGD SCOP SGD STRING Tair TaxID Unigene UniParc UniprotEntry WormBaseGeneID WormBaseSequenceName YPD AccessionNumber RefSeq TIGR UniprotAccession Disease Function Keyword Description SubcellularLocation Name Pubmed Formula Pvalue Score ProteinSequence Pattern STRINGScore STRINGScore_neighborhood STRINGScore_fusion STRINGScore_cooccurence STRINGScore_coexpression STRINGScore_experimental STRINGScore_db STRINGScore_textmining SequenceMap NucleotideSequence PDB TaxID_category TaxID_name GO_name - External Entity Relation Participant Attributes
cardinality detection_method GO KeggCode role
And here is the list of valid BIANA types:
- External Entity Types
protein DNA RNA mRNA tRNA rRNA CDS gene sRNA snRNA snoRNA structure pattern compound drug glycan enzyme relation ontology SCOPElement taxonomyElement PsiMiOboOntologyElement GOElement - External Entity Relation Types
interaction no_interaction reaction functional_association cluster homology pathway alignment complex regulation cooperation forward_reaction backward_reaction
In case, you need to annotate your data with some attribute or type that does not belong to the lists given above, you can use the following methods to introduce your attributes and types to BIANA . To add an;
- External Entity Type
- add_valid_external_entity_type( new_type )
- External Entity Relation
- add_valid_external_entity_relation_type( new_type )
- External Entity Attribute (Textual)
- add_valid_external_entity_attribute_type( new_attribute, data_type, ``eE identifier attribute'' )
- External Entity Attribute (Numeric)
- add_valid_external_entity_attribute_type( new_attribute, data_type, ``eE numeric attribute'' )
- External Entity Relation Attribute (Textual)
- add_valid_external_entity_attribute_type( new_attribute, data_type, ``eE identifier attribute'' )
- External Entity Relation Attribute (Numeric)
- add_valid_external_entity_attribute_type( new_attribute, data_type, ``eE numeric attribute'' )
- External Entity Relation Participant Attribute
- add_valid_external_entity_relation_participant_attribute_type( new_attribute, data_type )
6.6 Proposed unification protocol
List of external databases and the attributes (identifiers) proposed to be used in a unification protocol are given below.
| External Databases | Attributes (identifiers) |
|---|---|
| Uniprot, GeneBank, IPI, KeggGene, COG, String | ProteinSequence AND taxID |
| Uniprot, HGNC, HPRD, DIP, MPACT, Reactome, IPI, BioGrid, MINT, IntAct, String | UniprotAccession |
| Uniprot, String | UniprotEntry |
| Uniprot, HGNC, HPRD, DIP, String | GeneID |
| Uniprot, SCOP(promiscuous) | PDB |
7. Additional administration utilities
In this section, some additional administration commands are explained. Some of the commands are in the scripts folder of the application, and other are usual command line commands. Some of the commands are available for Windows and UNIX systems, and some others only for UNIX Systems.
7.1 BIANA database backup
In order to do a backup of the BIANA Database, it is only necessary to use the mysqldump\ utility provided with MySQL. Having a database dump is a good idea for the following reasons:
- Copying a database from one server to another without having to parse all databases again (easier, faster).
- Having a backup of the data used to perform specific experiments.
The command necessary to get the dump file and compress it is:
{parser_command}
\$> mysqldump --opt --user=USER --password=PASSWORD
--host=HOST BIANA_DATABASE_NAME | gzip -c > database_backup_X.sql.gz
* In Windows, probably the gzip program is not available. Skip it, and the compress the file with you compression program.
The commands necessary to put the data into a new database are:
1) Create a new mysql database:
{parser_command}
\$> mysql --user=USER --password=PASSWORD --host=HOST -e
"CREATE DATABASE DATABASE_NAME"
2) Insert the data into the database:
{parser_command}
\$> gunzip -c your_backup_mysql_file.sql.gz | mysql
--user=USER --password=PASSWORD --host=HOST "DATABASE_NAME"
* In Windows, probably the gunzip program is not available by default. You must uncompress the file using tools as WinZip and the execute the mysql command..
8. Glossary
- BIANA Database
- A repository of BIANA containing set of external databases and unified entries compiled from all available external databases based on specified unification protocols.
- External Database
- Any data source that contains biologic or chemical data that can be parsed by BIANA .
- External Entity
- Any entry found in any external database, such as a uniprot entry (a protein), a GenBank entry (a gene), an IntAct interaction (an interaction), a KEGG pathway or a PFAM alignment.
- External Entity Attribute
- Element associated to an external entity. External entities are characterized by several attributes such as database identifiers, descriptions, function, disease, ...Each external entity attribute has a distinct meaning. Each external entity is characterized by its associated attributes.
- External Entity Relation
- Any relation between two or more external entities.
- Hub User Entity
- User entity that has a number of connections in a relationships network higher than a given cutoff.
- Leaf User Entity
- User entity that only contains an edge in a relationship network.
- Linker User Entity
- Given a relationship graph between user entities, a user entity is considered linker only if it belongs to the path that links two or more seed user entities.
- Promiscuous external database
- External database whose external entities can belong to multiple User Entities in the same unification protocol.
- Unification Protocol
- Set of rules (unification protocol atoms) that determine how data in various data sources are combined (crossed). All unification protocol atoms are used in with union strategy (OR), i.e. rule1 OR rule2 OR rule3.
- Unification Protocol Atom
- Rule that determines how data in two external databases should be crossed. It is composed by two external databases (that can be the same or not) and one or more attributes. All attributes in an unification protocol atom are used with intersection strategy (AND). For example, external entities from external databases 1 and 2 are going to be considered equivalent if they share sequence similarity and taxonomy id.
- User Entity
- Set of external entities considered as equivalent as the result of applying a unification protocol. Each user entity has a unique identifier for each unification protocol. An external entity can belong only to one user entity (if the external database is not promiscuous), but a user entity can be composed by several external entities.
- User Entity Level
- Maximum number of connections between any seed node and any other non-seed node.
- User Entity Set
- Set of user entities result of a user experiment. User entity set contains the seed user entities obtained when creating the set, and all the user entities obtained when creating the network. User entity set is characterized by the user entities it contains, as well as the levels of their nodes, tags assigned to nodes and relations, groups of nodes by some criteria, etc.
- Seed User Entity
- User entity used in the first step of network creation (user entities belonging to level 0).
9. Frequently Asked Questions (FAQs)
- Installation
- How do I check whether BIANA python package is installed properly?
- Execute python interpreter and try to import the package as below. If you interpreter prompts ``BIANA>'' string, BIANA is installed properly.
{bianacommand} \$>python >>> import biana BIANA>If not, revisit the installation instructions and make sure that BIANA python package is installed properly.
- Execute python interpreter and try to import the package as below. If you interpreter prompts ``BIANA>'' string, BIANA is installed properly.
- What to do if Cytoscape gives BIANA package import error while starting BIANA plugin?
- Make sure that you configured PYTHONPATH environment variable to include directory where BIANA python package is installed.
- Restart your computer (to make sure that Cytoscape sees the changes you made to PYTHONPATH).
- What should I do if I get ``can not connect to MySQL database'' error?
- Check that MySQL server is running properly and (re)start it if necessary.
- Check that database host and user information you provide is correct.
- If you are MySQL server in your local host try using ``127.0.0.1'' instead of ``localhost'' as host name.
- How do I check whether BIANA python package is installed properly?
- Database population
- Is it normal that populating a BIANA Databasetakes more time than expected?
- BIANA Databasepopulation time will depend on multiple factors that can produce differences in parsing between different computers: parsed database, disk free space, disk access speed, network speed if MySQL server is in other computer... BIANA Databasecan have two distinct states: Running and parsing. When parsing, BIANA Databasestate is in ``parsing'' mode, and when starting a BIANA Session it changes automatically to ``running'' mode.
- Why does BIANA not recognize, a new parser I have created?
- Make sure you copied your new parser into BIANA _Installation_ path/bianaParser. Then, BIANA should recognize the parser and it will appear in the graphical interface as well. If you are not sure where BIANA was installed, execute python interpreter and try the following.
{bianacommand} \$> python >>> import biana BIANA> biana.__path__
- Make sure you copied your new parser into BIANA _Installation_ path/bianaParser. Then, BIANA should recognize the parser and it will appear in the graphical interface as well. If you are not sure where BIANA was installed, execute python interpreter and try the following.
- Is it normal that populating a BIANA Databasetakes more time than expected?
- Data Unification
- What is the best unification protocol to use, do you have any suggested unification protocols?
- Create & use a unification protocol that suits best to your needs (specific to your problem). You may want to check 6.6 proposed unification protocol section to have some ideas.
- What is the best unification protocol to use, do you have any suggested unification protocols?
- BIANA Execution
- Why does the message ``Optimizing database...'' appear during a long time, when I start a BIANA Session?
- This message only appears the first time you start a Session in a BIANA Databaseafter adding a new external database in it. This process creates all necessary indices in the database to increase performance while running, and it is done after populating database in order to increase parsing performance. Depending on the BIANA Database size, this process can take from few seconds to a couple of hours.
- If it takes too long, check your disk space where BIANA Database is stored is not full!
- I do not have any entries when I create a new user entity set, what could be the reason?
- If you created a unification protocol using only promiscuous databases, it is normal that you do not have any user entities. Data coming from promiscuous databases added to (multiple) user entities that contain at least one entry coming from a non-promiscuous entry.
- There may not be any entry associated with your query, try to refine the attributes and values you have used.
- Is it normal that creating networks takes too much time?
- Huge and very connected networks can take some time. Be patient. Be sure you are doing the network you want to the correct level. If you don't want interactions between elements at the last level, don't add them!
- If you are using the graphical interface as a Cytoscape Plugin, it usually slows down the process significantly. Execute the same process by command line. A trick is to create the set with Cytoscape, start the network at level 0 without relations at the last level, then save the commands into a file, then edit manually the file to set the correct level and finally execute the script.
- Database connection has been lost when using BIANA Cytoscape plugin. Should I restart the plugin?
- MySQL server usually closes connection after some time the connection has not been used (this time will depend on your MySQL server configuration). It is not necessary to restart the plugin, right-click in the Biana Session and select the option ``Reconnect database''.
- When I execute BIANA Cytoscape plugin, it is very slow or Cytoscape exits suddenly.
- When executing BIANA as a Cytoscape plugin, it consumes more time and memory than executing it as a command line application. You have different options:
- By default, cytoscape.sh uses a maximum memory limit of 512Mb. If the program exceeds it, it will automatically exit without saving anything. You can increase this memory limit by modifying the parameter -Xmx512M when executing cytoscape.
- If you are creating a huge network and you are not interested in visualizing it but only in getting the data, use BIANA scripts: it will be faster and it will require less computer resources. You can use the following trick to create the script you are interested in:
- Run BIANA Cytoscape plugin, create the network at level 0 and perform all the operations you are interested in.
- In the Session Pop-up Menu (right-click on BIANA Session), select the option ``Save commands history''.
- Modify the saved script by changing all the parameters you want and execute it from command line.
- When executing BIANA as a Cytoscape plugin, it consumes more time and memory than executing it as a command line application. You have different options:
- Why does the message ``Optimizing database...'' appear during a long time, when I start a BIANA Session?
2009-11-03