
|

|

|
| Information on 1g9g |
| PDB: 1g9g Compound: cellulase cel48f Classification: HYDROLASE Entry date in PDB: 2003-06-24 Resolution [Å]: 1.90 R-Factor: 0.165 |
CHAIN: A SWISS-PROT/TREMBL:
P37698
KEYWORD: 3D-structure Cellulose degradation Glycosidase Hydrolase Repeat Signal EC: 3.2.1.4 SCOP: a.102.1.2 All alpha proteins alpha/alpha toroid Six-hairpin glycosidases Cellulases catalytic domain Processive endocellulase CelF (Cel48F) Clostridium cellulolyticum GO: hydrolase activity, hydrolyzing O-glycosyl compounds carbohydrate metabolism polysaccharide catabolism |
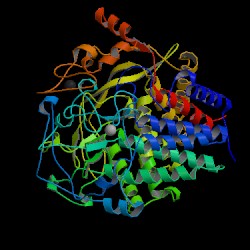
Image Source: PDB |
| Stored Loops of 1g9g |
Loops in ArchDB40 clusters
1g9g_A_515 - HE => SUBCLASS : 0.1.7
1g9g_A_515 - HE => SUBCLASS : 0.1.12
1g9g_A_432 - HE => SUBCLASS : 1.2.1
1g9g_A_391 - HA => SUBCLASS : 2.1.1
1g9g_A_563 - HA => SUBCLASS : 3.1.1
1g9g_A_450 - HA => SUBCLASS : 5.1.4
1g9g_A_301 - HA => SUBCLASS : 5.11.1
1g9g_A_485 - EH => SUBCLASS : 2.4.2
1g9g_A_194 - AR => SUBCLASS : 1.1.21
1g9g_A_495 - HH => SUBCLASS : 2.2.1
1g9g_A_413 - HH => SUBCLASS : 2.2.3
1g9g_A_413 - HH => SUBCLASS : 2.2.5
1g9g_A_54 - HH => SUBCLASS : 2.2.6
1g9g_A_413 - HH => SUBCLASS : 2.14.1
1g9g_A_229 - HH => SUBCLASS : 3.2.14
Loops in ArchDB95 clusters
1g9g_A_485 - EH => SUBCLASS : 2.3.1
1g9g_A_485 - EH => SUBCLASS : 2.3.3
1g9g_A_194 - AR => SUBCLASS : 1.2.2
1g9g_A_495 - HH => SUBCLASS : 2.2.1
1g9g_A_413 - HH => SUBCLASS : 2.2.3
1g9g_A_54 - HH => SUBCLASS : 2.2.6
1g9g_A_515 - HE => SUBCLASS : 0.1.16
1g9g_A_432 - HE => SUBCLASS : 1.3.1
1g9g_A_391 - HA => SUBCLASS : 2.1.1
1g9g_A_563 - HA => SUBCLASS : 3.1.1
1g9g_A_450 - HA => SUBCLASS : 5.4.2
1g9g_A_301 - HA => SUBCLASS : 5.10.1
Loops in ArchDB-EC clusters
1g9g_A_54 - HH => SUBCLASS : 2.2.17
Loops not clustered in ArchDB
1g9g_A_537 - AR
1g9g_A_197 - AR
1g9g_A_213 - AR
1g9g_A_311 - AR
1g9g_A_373 - AR
1g9g_A_194 - AR
1g9g_A_197 - AR
1g9g_A_281 - AR
1g9g_A_213 - AR
1g9g_A_281 - AR
1g9g_A_311 - AR
1g9g_A_373 - AR
1g9g_A_537 - AR
1g9g_A_541 - EH
1g9g_A_222 - EH
1g9g_A_51 - EH
1g9g_A_604 - EH
1g9g_A_570 - EH
1g9g_A_395 - EH
1g9g_A_123 - EH
1g9g_A_318 - EH
1g9g_A_277 - HA
1g9g_A_301 - HA
1g9g_A_563 - HA
1g9g_A_391 - HA
1g9g_A_450 - HA
1g9g_A_457 - HA
1g9g_A_533 - HA
1g9g_A_175 - HA
1g9g_A_155 - HA
1g9g_A_175 - HA
1g9g_A_277 - HA
1g9g_A_457 - HA
1g9g_A_155 - HA
1g9g_A_533 - HA
1g9g_A_7 - HE
1g9g_A_549 - HE
1g9g_A_345 - HE
1g9g_A_254 - HE
1g9g_A_137 - HE
1g9g_A_97 - HE
1g9g_A_590 - HE
1g9g_A_326 - HH
1g9g_A_75 - HH
| Homologous structures to 1g9g classified in ArchDB |
1f9d A - percentage of sequence identity: 99
1f9o A - percentage of sequence identity: 100
1fae A - percentage of sequence identity: 100
1fbo A - percentage of sequence identity: 100
1fbw A - percentage of sequence identity: 99
1fce - - percentage of sequence identity: 100
1g9j A - percentage of sequence identity: 99
1l1y B - percentage of sequence identity: 60
1l1y C - percentage of sequence identity: 60
1l1y D - percentage of sequence identity: 60
1l1y E - percentage of sequence identity: 60
1l1y F - percentage of sequence identity: 60
1l2a A - percentage of sequence identity: 60
1l2a B - percentage of sequence identity: 60
1l2a C - percentage of sequence identity: 60
1l2a D - percentage of sequence identity: 60
1l2a E - percentage of sequence identity: 60
1l2a F - percentage of sequence identity: 60