
|

|

|
| Information on 1id2 |
| PDB: 1id2 Compound: amicyanin Classification: ELECTRON TRANSPORT Entry date in PDB: 2001-04-11 Resolution [Å]: 2.15 R-Factor: 0.174 |
CHAIN: A SWISS-PROT/TREMBL:
P22365
KEYWORD: 3D-structure Copper Electron transport Periplasmic Pyrrolidone carboxylic acid Signal SCOP: b.6.1.1 All beta proteins Cupredoxin-like Cupredoxins Plastocyanin/azurin-like Amicyanin Paracoccus versutus (Thiobacillus versutus) GO: copper ion binding electron transporter activity electron transport CHAIN: B SWISS-PROT/TREMBL:
P22365
KEYWORD: 3D-structure Copper Electron transport Periplasmic Pyrrolidone carboxylic acid Signal SCOP: b.6.1.1 All beta proteins Cupredoxin-like Cupredoxins Plastocyanin/azurin-like Amicyanin Paracoccus versutus (Thiobacillus versutus) GO: copper ion binding electron transporter activity electron transport CHAIN: C SWISS-PROT/TREMBL:
P22365
KEYWORD: 3D-structure Copper Electron transport Periplasmic Pyrrolidone carboxylic acid Signal SCOP: b.6.1.1 All beta proteins Cupredoxin-like Cupredoxins Plastocyanin/azurin-like Amicyanin Paracoccus versutus (Thiobacillus versutus) GO: copper ion binding electron transporter activity electron transport |
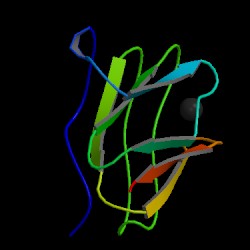
Image Source: PDB |
| Stored Loops of 1id2 |
Loops in ArchDB95 clusters
1id2_A_77 - AR => SUBCLASS : 3.1.2
1id2_A_30 - AR => SUBCLASS : 3.4.2
1id2_A_77 - AR => SUBCLASS : 4.3.1
1id2_A_22 - HA => SUBCLASS : 2.1.1
1id2_A_87 - HA => SUBCLASS : 6.3.1
Loops not clustered in ArchDB
1id2_A_4 - AR
1id2_A_12 - AR
1id2_A_35 - AR
1id2_A_43 - AR
1id2_A_68 - AR
1id2_A_57 - HA
| Homologous structures to 1id2 classified in ArchDB |
1aac - - percentage of sequence identity: 62
1aaj - - percentage of sequence identity: 62
1aan - - percentage of sequence identity: 62
1bxa A - percentage of sequence identity: 62
1id2 A - percentage of sequence identity: 100
1id2 B - percentage of sequence identity: 100
1id2 C - percentage of sequence identity: 100
1mda A - percentage of sequence identity: 62
1mda B - percentage of sequence identity: 62
1mg2 C - percentage of sequence identity: 62
1mg2 G - percentage of sequence identity: 62
1mg2 K - percentage of sequence identity: 62
1mg2 O - percentage of sequence identity: 62
1mg3 C - percentage of sequence identity: 62
1mg3 G - percentage of sequence identity: 62
1mg3 K - percentage of sequence identity: 62
1mg3 O - percentage of sequence identity: 62
1sf3 A - percentage of sequence identity: 61
1sf5 A - percentage of sequence identity: 61
1sfd A - percentage of sequence identity: 61
1sfd B - percentage of sequence identity: 61
1sfh A - percentage of sequence identity: 61
1sfh B - percentage of sequence identity: 61
1t5k A - percentage of sequence identity: 62
1t5k B - percentage of sequence identity: 62
1t5k C - percentage of sequence identity: 62
1t5k D - percentage of sequence identity: 62
2mta A - percentage of sequence identity: 62
2rac A - percentage of sequence identity: 62