
|

|

|
| Information on 1gvj |
| PDB: 1gvj Compound: c-ets-1 protein Classification: TRANSCRIPTION Entry date in PDB: 2004-02-06 Resolution [Å]: 1.53 R-Factor: 0.208 |
CHAIN: A SWISS-PROT/TREMBL:
P14921
KEYWORD: 3D-structure Alternative splicing DNA-binding Nuclear protein Phosphorylation Proto-oncogene Transcription regulation 3D-structure Alternative splicing DNA-binding Nuclear protein Phosphorylation Proto-oncogene Transcription regulation SCOP: a.4.5.21 All alpha proteins DNA/RNA-binding 3-helical bundle "Winged helix" DNA-binding domain ets domain ETS-1 transcription factor, residues 331-440 Human (Homo sapiens) GO: nucleus DNA binding transcription factor activity regulation of transcription, DNA-dependent CHAIN: B SWISS-PROT/TREMBL:
P14921
KEYWORD: 3D-structure Alternative splicing DNA-binding Nuclear protein Phosphorylation Proto-oncogene Transcription regulation 3D-structure Alternative splicing DNA-binding Nuclear protein Phosphorylation Proto-oncogene Transcription regulation SCOP: a.4.5.21 All alpha proteins DNA/RNA-binding 3-helical bundle "Winged helix" DNA-binding domain ets domain ETS-1 transcription factor, residues 331-440 Human (Homo sapiens) GO: nucleus DNA binding transcription factor activity regulation of transcription, DNA-dependent |
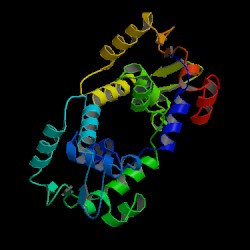
Image Source: PDB |
| Stored Loops of 1gvj |
Loops in ArchDB40 clusters
1gvj_A_362 - EH => SUBCLASS : 3.7.1
1gvj_A_418 - HH => SUBCLASS : 3.1.2
Loops in ArchDB95 clusters
1gvj_A_362 - EH => SUBCLASS : 3.3.3
1gvj_A_418 - HH => SUBCLASS : 3.4.2
1gvj_A_368 - HH => SUBCLASS : 6.16.1
1gvj_A_386 - HE => SUBCLASS : 2.4.1
1gvj_A_337 - HE => SUBCLASS : 7.12.1
1gvj_A_402 - HA => SUBCLASS : 6.8.2
Loops not clustered in ArchDB
1gvj_A_411 - EH
1gvj_A_355 - HA
1gvj_A_304 - HH
1gvj_A_323 - HH
| Homologous structures to 1gvj classified in ArchDB |
1awc A - percentage of sequence identity: 64
1bc8 C - percentage of sequence identity: 60
1dux C - percentage of sequence identity: 63
1dux F - percentage of sequence identity: 63
1fli A - percentage of sequence identity: 70
1gvj A - percentage of sequence identity: 100
1gvj B - percentage of sequence identity: 100
1k78 B - percentage of sequence identity: 100
1k78 F - percentage of sequence identity: 100
1k79 A - percentage of sequence identity: 100
1k79 D - percentage of sequence identity: 100
1k7a A - percentage of sequence identity: 100
1k7a D - percentage of sequence identity: 100
1md0 A - percentage of sequence identity: 100
1md0 B - percentage of sequence identity: 100
1mdm B - percentage of sequence identity: 100
1r36 A - percentage of sequence identity: 99
2stt A - percentage of sequence identity: 100
2stw A - percentage of sequence identity: 100