Thrombin is a trypsin-like serine proteinase that plays a crucial role in the processes of thrombosis and haemostasis.
The first structure of thrombin, solved in 1989, revealed an overall protein fold very similar to other members of the chymotrypsin family of proteases, including the characteristic catalytic triad of histidine 57, aspartate 102, and serine 195 (chymotrypsin template numbering is used throughout) in an active site cleft.The structure of thrombin differs from that of its simpler cousins by the presence of extension loops that line the walls of the active site cleft, the 60- and Gamma-loops, and 2 regions of positive electrostatic potential known as anionbinding exosites I and II.
Thrombin is generated from its zymogen prothrombin through proteolytic cleavage at 2 sites (arginines 271 and 320) by factor Xa. The resulting 37-kDa serine protease thrombin is no longer associated to its Gla and 2 Kringle domains and is thus free to diffuse away from the surface on which it was generated, and its special structural features are used to recognize a broad spectrum of substrates.
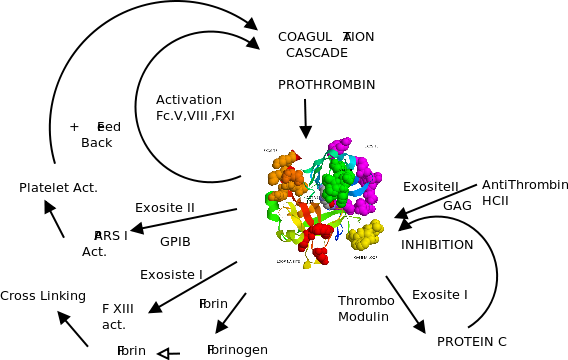
Thrombin is composed of two polypeptide chains of 36 (A chain) and 259 (B chain) residues,after the cleavage by factor Xa at residue 49. Both chains are covalently linked through a disulfide bond between residues C1 and C122.The four disulfide bridges of thrombin are homologous to those founds in chymotrypsins.
1.Cartoons representation thrombin(Heavy and Light(pink)) and sulfide bonds.
Thrombin like trypsin cleaves peptide bonds involving arginine residues however thrombin is more selective and only cleaves peptide bonds in a limited substrates.
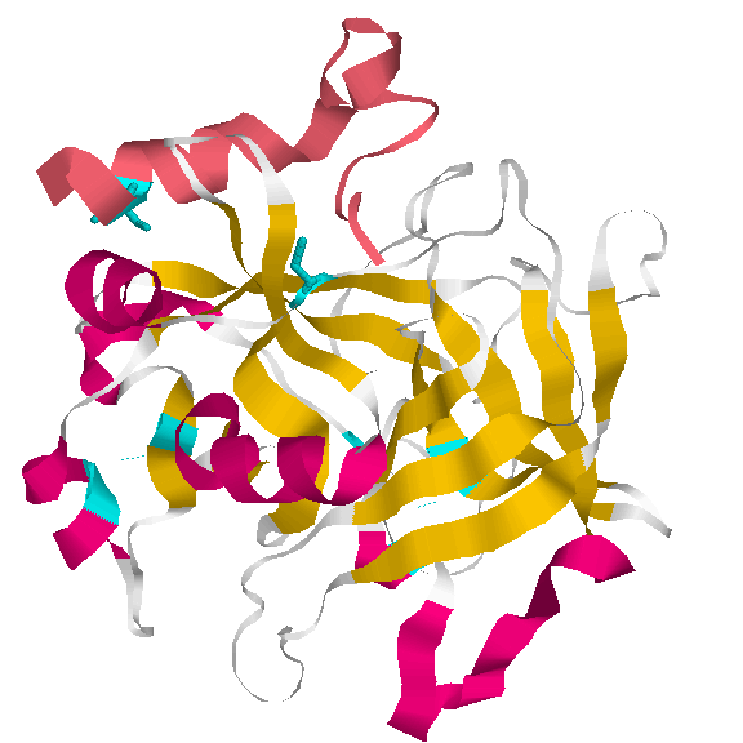
A common feature of trypsin-like protease is the presence of a serine residue(S195) in the active site cleft. In thrombin,S195,in conjunction with H57(Histidine) and D189(Aspartic acid) forms the active center.
sp|P00760|TRY1_BOVIN -----MKTFIFLALLGAAVAFP----------VDD-----------DDKI 24
P00735|318-625|Thrombin_BOVIN TSEDHFQPFFNEKTFGAGEADCGLRPLFEKKQVQDQTEKELFESYIEGRI 50
::.*: :**. * *:* :.:*
sp|P00760|TRY1_BOVIN VGGYTCGANTVPYQVSLN---SGYHFCGGSLINSQWVVSAAHCYKS---- 67
P00735|318-625|Thrombin_BOVIN VEGQDAEVGLSPWQVMLFRKSPQELLCGASLISDRWVLTAAHCLLYPPWD 100
* * . .. *:** * . :**.***..:**::****
sp|P00760|TRY1_BOVIN ------GIQVRLGEDNINVVEGNEQFISAS-KSIVHPSYN-SNTLNNDIM 109
P00735|318-625|Thrombin_BOVIN KNFTVDDLLVRIGKHSRTRYERKVEKISMLDKIYIHPRYNWKENLDRDIA 150
.: **:*:.. . * : : ** * :** ** .:.*:.**
sp|P00760|TRY1_BOVIN LIKLKSAASLNSRVASISLP-----TSCASAGTQCLISGWGN-----TKS 149
P00735|318-625|Thrombin_BOVIN LLKLKRPIELSDYIHPVCLPDKQTAAKLLHAGFKGRVTGWGNRRETWTTS 200
*:*** . .*.. : .:.** :. ** : ::**** *.*
sp|P00760|TRY1_BOVIN SGTSYPDVLKCLKAPILSDSSCKSAYPGQITSNMFCAGYLEG-GK--DSC 196
P00735|318-625|Thrombin_BOVIN VAEVQPSVLQVVNLPLVERPVCKASTRIRITDNMFCAGYKPGEGKRGDAC 250
. *.**: :: *::. . **:: :**.******* * ** *:*
sp|P00760|TRY1_BOVIN QGDSGGPVVCSG------KLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK 240
P00735|318-625|Thrombin_BOVIN EGDSGGPFVMKSPYNNRWYQMGIVSWGEGCDRDGKYGFYTHVFRLKKWIQ 300
:******.* .. ******.** :..* *.**:* . .**:
sp|P00760|TRY1_BOVIN QTIASN-- 246
P00735|318-625|Thrombin_BOVIN KVIDRLGS 308
:.*
Thrombin Site active.
Histidine(green),Aspartic acid(red),Serine(yellow)
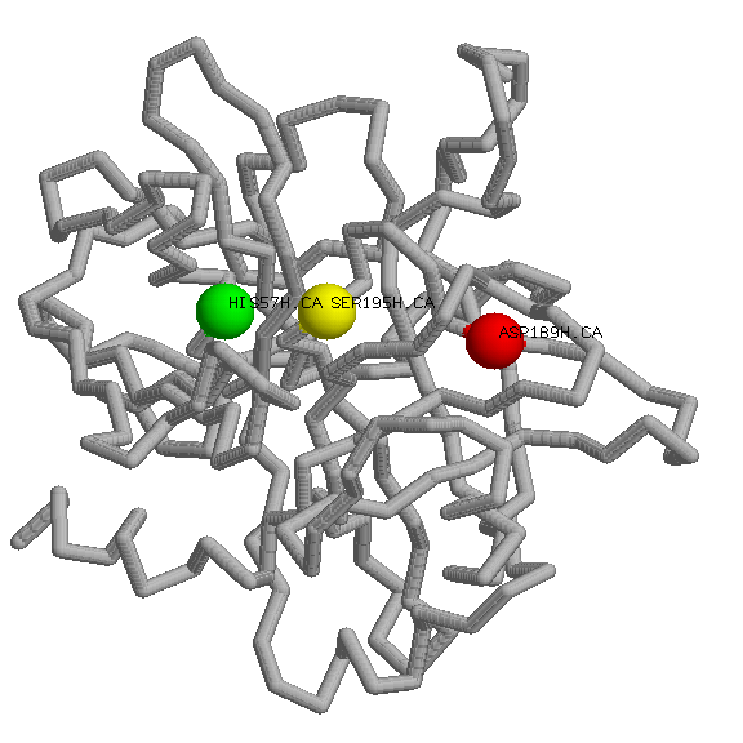
Thrombin substrate specificity is not determined primarily by the active site, rather by a inaccesible active site due two striking loop insertions and also two patches of positive surface potential that mediate intermolecular interactions: the anions binding exosites. Thrombin specificity can thus be understood as a competition for three sites: the active site, and exosites I and II.
A.ANION BINDING SITES
Exosites are surface sites on proteinases physically separated from the active site residues that determine the primary S1-S4 peptide substrate and inhibitor specificity. Thrombin has two anion-binding exosites (I and II) that are made up of distinct clusters of surface exposed basic residues. These charged patches interact specifically with negatively charged regions on thrombin's cofactors and substrates.
Exosite I: Is located nearly 20 Anstroms distal to the active site. Composed of insertion loops 30-40 and 70-80,mediates binding of fibrinogen, fibrin, PAR-1 and -4 substrate recognition,etc.
Exosite II:Is located 10 Anstroms to the west of Asp-102 in the catalytic triad and is formed by a cluster of residues on and about the C-terminal helix(R93, K236, K240, R101, and R233). It mediates different interactions specifically, including heparin, a specific monoclonal antibody, and the fragment 2 domain of ProT. A variety of inhibitors from snake venoms and the saliva of hematophagous organisms also bind to thrombin exosites.
B.INSERTION LOOPS
The insertion loops forms like canyon a structure with the active site at bottom. They restrict access to the catalytic site of thrombin to proteins with long, flexible substrate loops, or to proteins with complementary surfaces which mediate contact with the loops.
60-Loop:
The 60-loop is composed of Leu60, Tyr60a, Pro60b, Pro60c, Trp60d, Asp60e, Lys60f, Asn60g, Phe60h, and Thr60i, with Asn60g glycosylated. From the sequence we can see that is hydrophobic in nature, with the two consecutive prolines serving to rigidify. The 60-loop, thus provides a rigid, hydrophobic cap over the active site and mediates contacts with the hydrophobic substrate residues N-terminal to the scissile bond.
Thrombin: SER45 LEU46 ILE47 SER48 ASP49 ARG50 TRP51 VAL52 LEU53 THR54
ALA55 ALA56 HIS57 CYS58 LEU59 LEU60 TYR60 PRO60 PRO60 TRP60
ASP60 LYS60 ASN60 PHE60 THR60 VAL61 ASP62 ASP63 LEU64 LEU65
Chimiotrypsin: ALA56 HIS57 CYS58 GLY59 VAL60 THR61 THR62 SER63 ASP64 VAL65
VAL66 VAL67 ALA68 GLY69 GLU70 PHE71 ASP72 GLN73 GLY74 SER75
Gamma Loop:
The Gamma Loop is composed of Thr147, Trp147a, Thr147b, Ala147c, Asn147d, and Val147f.It's more hydrophilic and flexible in nature It is often incompletely modeled in crystal structures due to its inherent mobility, but, as it is adjacent to the active site cleft of thrombin, it can contact substrate residues C-terminal to the scissile bond, and can make contacts with the body of substrate proteins.
C.THE NA+-DEPENDENT ALLOSTERY
The thrombin catalytic site and the affinity of exosite I ligands are allosterically regulated by binding of Na+. The residues energetically linked to Na+-induced allostery are D189, E217, D222, and Y225. Na+ binding switches thrombin from a 'slow' to a 'fast form' with higher specificity (kcat/Km) for the procoagulant substrates, fibrinogen, and PAR-1, greater reactivity toward antithrombin, and increased peptide substrate activity The Na+-free, slow form has higher relative specificity in protein C activation. Thus, Na+ regulates the procoagulant and anticoagulant activities of thrombin.
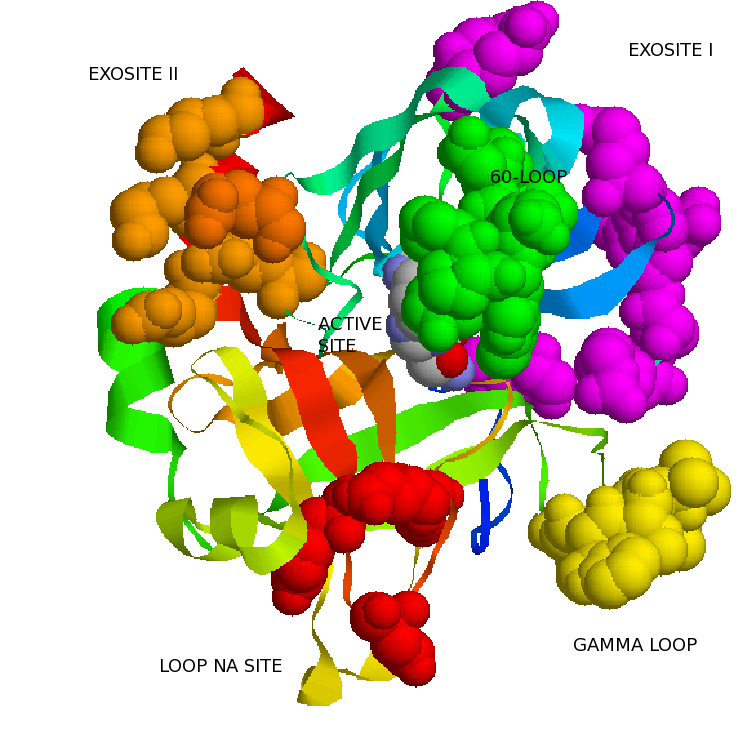
Following initiation of coagulation as part of the hemostatic response to injury, thrombin is generated from its inactive precursor prothrombin by factor Xa as part of the prothrombinase complex.
Thrombin then acts in different ways orchestratedby different interactions with substrates and cofactors.It is a multifuncional protein. Thrombin acts as procoagulant but also as anticoagulant, but also as anti-Fibrinolytic anti-inflamatory. It seems as thrombin goes throught different functions,stages like in a lyfe-cycle.
We require knowledge of its structure to determine how it interacts with its substrates, and the role of any cofactors for its interaction with substrates.
Biochemical and mutagenesis studies have established that thrombin recognition of substrates invariably involves engagement of the active site cleft and either one or both of the exosite.
In addition to direct interaction with substrates, thrombin specificity is also regulated through interaction with several cofactors. Cofactor binding regulates thrombin activity toward various substrates by mechanisms such as localization, competition for exosite binding, and allostery.
Cofactor
GpIb-alpha
Fibrin
Thrombomodulin
Glycosaminoglycan
Na+
|
Substrate and Fold activation
PAR-1; GpV; Factor XI
Factor XIII
PC;TAFI
AT;HCII
Fibrinogen
|
The major procoagulant functions(i.e cleavage of fibrinogen,FV and FVIII) of thrombin occur without cofactors. All of these reactions involve extended interactions of both exosite I and II on thrombin with the target substrate. In addition,the Na+-binding loop and the active site are implied.
Sodium
The binding site for Na+ was first identified in 1995 by Di Cera et al. When bound to Na+, thrombin exists in a prothrombotic state capable of efficient cleavage of procoagulant substrates like fibrinogen and PAR-1.
In the absence of Na+, thrombin displays a generally reduced catalytic efficiency for these and other substrates yet maintains the ability to effectively activate protein C in the presence of TM.
GpIb-alpha
GpIb-alpha is a heavily O-glycosylated transmembrane protein that has been hypothesized to be the high-affinity binding site for thrombin on the platelet surface.It acts as a cofactor by enhancing thrombin cleavage of the platelet receptor PAR-1, resulting in platelet activation.
The binding site for GpIb alpha on thrombin has been localized to exosite II, with mutations in this exosite displaying a marked decrease in affinity for GpIb alpha,whereas similar mutations in exosite I of thrombin showed little or no effect.Exosite II residue arginine 233 is critical for the interaction
Fibrin
Fibrin polymers can act as a cofactor for the thrombin activation of the transglutaminase factor XIII, which cross-links the fibrin to strengthen the clot.Thrombin interacts with fibrin via exosite I residues phenylalanine 34, serine 36a, leucine 76, arginine 77a, isoleucine 82, and lysine 110. Although the interface juxtaposes complementary charged regions on thrombin and fibrin, hydrophobic interactions provide the bulk of the binding energy.
Thrombomodulin
Thrombomodulin is expressed on the surface of endothelial cells and binds thrombin with high affinity. Binding to TM alters thrombin specificity from procoagulant substrates toward activation of the anticoagulant protease protein C.8 Thrombin interacts with TM primarily through exosite I, yet exosite II can also be involved. The majority of binding energy is supplied by hydrophobic contact.
Glycosaminoglycan(GAGs)
GAGs, such as heparan sulfate, chondroitin sulfate, and dermatan sulfate, are long-chain sulfated polysaccharides attached to proteoglycans that coat the surface of cells lining the vascular and extravascular spaces. Heparin, the widely used anticoagulant drug, is a relatively small and uniform GAG secreted by mast cells. GAGs bind to thrombin and have been shown to significantly accelerate the inhibition of thrombin by serpins,antithrombin (AT) and heparin cofactor II (HCII), through a bridging mechanism.
Mutagenesis studies have identified the heparin binding site of thrombin as exosite II and includes (in order of importance) arginine 93, lysine 236, lysine 240, arginine 101, and arginine 233.