A typical genome contains 2-4% of genes encoding for proteolytic enzymes,proteinases. Proteinases are enzymes that catalyse hydrolyse of peptide bonds in different substrates. In their absence reactions proceed at kinetically insignificant rates incompatible with living.
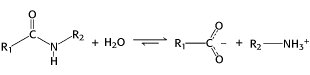
Proteases cleave proteins by a hydrolysis reaction,the addition of a molecule of water to a peptide bond.
Proteinases can be classified into 4 families based on functional criterion: Serine,Cysteine,aspartic or metallo proteinases. What differentitate the differents families is the nature of the most prominent functional group in the active site.
Barrett and coworkers have devised a classification scheme based on statistically significant similarities in sequence and structure of all known proteolytic enzymes and term this database MEROPS.
UniProt - Swiss-Prot Classification
Each proteinase type can be classified into different families and clans.
Members of the same clan are proteases that have evolved from a common ancestor and share a common protein fold.
Proteases in the same family are related based on the sequence homology of their amino acid sequences.
Most, but not all, clans consist of one active site arrangement. The prefix 'S' is used for clans formed entirely from families which are serine peptidases, whereas 'P' is used for clans in which the homologous families are of several catalytic types (e.g., serine and cysteine).Another remarkable point is that since some members of the same clan can use different active site architectures, this indicates that the tertiary structure is not always related to the active site configuration.
Over one third of all known proteolytic enzymes are serine proteases, which during evolution emerged as the most abundant and functionally group.The family name stems from the nucleophilic Ser in the enzyme active site. Serine proteases are grouped into 13 clans and 40 families.
Serine proteases are widely distributed in nature and found in all kingdoms of cellular life as well as many viral genomes. However, significant differences exist in the distribution of each clan across species. For example, clan PA proteases are highly represented in eukaryotes, but rare constituents of prokaryotic and plant genomes.
All serine proteinases have Serine in their active site,but the configuration of the active site can vary between them.
At least four distinct protein folds as illustrated by trypsin, subtilisin, prolyl oligopeptidase, and ClpP peptidase utilize the Asp-His-Ser catalytic triad in identical configuration to catalyze hydrolysis of peptide bonds. One difference between them is that the triad is ordered in a different way.In chymotripsin clan is ordered HDS,in subtilisin clan DHS and in carboxypeptidase clan SDH.This shows that the linear arrangements of the catalytic residues commonly reflect clan relationships.
On the other hand,many serine proteases employ a simpler dyad mechanism where Lys or His is paired with the catalytic Ser.
Other serine proteases mediate catalysis via novel triads of residues, such as a pair of His residues combined with the nucleophilic Ser.
The proteases with a Ser/His/Asp triad fall within four of these clans (PA, SB, SC, and SK), while the Ser/Lys proteases fall within five clans (SE, SF, SJ, SK, and SR).This shows that different ancestors can converge on the same Ser/His/Asp or Ser/Lys mechanism.
In Humans four protease families by themselves account for over 40% of all proteolytic enzymes:
-
Ubiq-uitin-specific proteases responsible for regulated intracellular protein turnover.
-
Adamalysins. controlling growth factors and integrin function.
-
Prolyl oligopeptidases.
-
trypsin-like.
Of 699 proteases in man, 178 are serine proteases and 138 of them belong to the S1 protease family.
Abundance of S1 proteases suggests the protein fold presents a selective advantage relative to other proteases. The trypsin fold of the S1 protease family (Fig. 1) nestles catalytic efficiency, substrate selectivity, and multiple levels of regulation in a scaffold that is readily associated with other auxiliary protein modules.
Clan PA proteases bearing the trypsin fold are the largest family of serine proteases.
Digestive enzymes such as trypsin and chymotrypsin cleave polypeptide chains at positively charged (Arg/Lys) or large hydrophobic (Phe/Trp/Tyr)residues, respectively. Most clan PA proteases have trypsin-like substrate specificity and prefer Arg or Lys side chains at the P1 position of substrate.
Important biological processes rely on clan PA proteases, like blood coagulation and the immune response,which involve cascades of sequential zymogen activation. In both systems, the protease domain is combined with one or more domains,like kringle domains and others. These domains are present on the N-terminus as an extension of the propeptide segment of the protease and typically remain attached to the protease domain through a covalent disulphide bond.
The S1 family comprises the largest of all serine proteases.Evolution has produced a wealth of diversity within the S1 peptidase family over 2000 protease sequences have been identified to date within S1 peptidases).Evolutionary trees constructed from only a short 50 amino acid stretch of residues in the C-terminal region of the protease that encompasses the active site of the enzyme are an equally informative evolutionary tree as one that incorporates the full protease domain and auxiliary domains.
