Serine proteinases are one of the four functional groups of the proteinases enzymes. They are widely distributed in the nature, and can be found in both eukariotyc as well as prokaryotic organisms. The key characterisic of serine proteinases consists of having a serine aminoacid in the active site.
The serine proteinases have been extensively studied, both by kinetic methods in solution, as well as by x-ray structural studies. Thanks to these study methodologies, the associate reaction mechanism has been stated. Following, in the next section, the serine proteinase catalysis is described.
In general, proteinases describe a very extensive group of enzymes that catalyse the hydrolysis of covalent peptidic bonds. More specifically, the catalytic mechanism of serine protreinases is a chemical reaction in which a peptide bond of a polypeptide chain is cleaved, producing two newer smaller peptides.
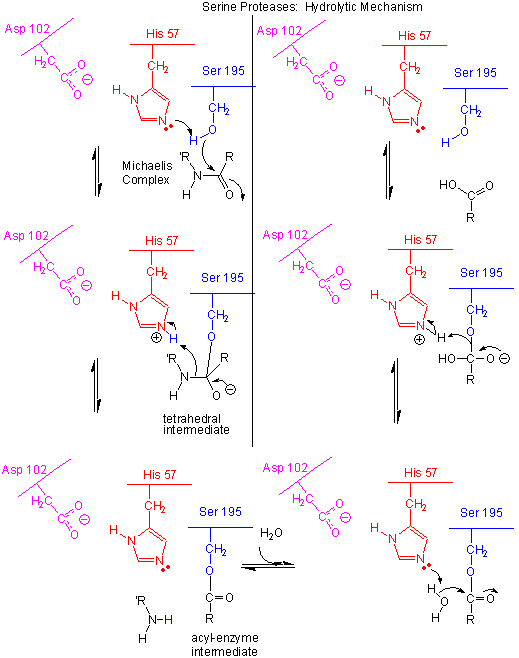
The serine proteinase catalysis is based on a nucleophilic attack to scissile bond of target polypeptide by a serine (Ser) aminoacid, contained in the active site of the protein. Moreover, this nucleophilic property of the serine (ser) is improved by the presence of the aminoacid histidine (His), which accepts a proton donated by a third aminoacid of the active site: the aspartate (Asp). These three aminoacids: serine (Ser), histine (His), and aspartate (Asp), are located very near one another near the heart of the enzyme, and play an essential role in the cleaving ability of the serine proteinases. They built the catalytic triad.
In a more detailed way, the serine proteinase catalysis is composed by a set of ordered steps. The peptide cleavage can be seen as a ping-pong catalysis process. Firstly, a substrate binds (in this case, the polypeptide being cleaved), then a product is released: the N-terminus half of the peptide. Secondly, another substrate binds (in this case, water), and finally, another product is released: the C-terminus half of the polypeptide binded at the beging. Following each sub-process is presented:
-
The deprotonated aminoacid histine (His) acts as a general base to attract a proton from serine (Ser). This enhances the nucleophilicty, and causes an attack to the electrophilic C of the amide, creating the oxyanion tetrahedral intermediate. The histine aminoacid (His), which has acquired positive charge, is stabilized by the aspartate (Asp) aminoacid.
-
The oxyanion hole collapses back to form a double bond between the O and the original carbonyl C, with the amine product as the leaving group. The protonated histine (His) acts as a general acid donating a proton to the amine leaving group, regenerating the unprotonated histine (His).
-
The mechanism repeats itself only now with water as the nucleophile, which attacks the acyl-enzyme intermediate, to form the tetrahedral intermediate.
-
The intermediate collapses again, releasing the C-terminus as the leaving group which gets reprotonated by the histine (His). This regenerates both the histine (His) and the serine (Ser) to the normal protonation state. Finally, the serine proteianse is ready for another catalytic round of activity.
In the following section, the common structural features of serine proteinases are presented.
The serine protreinases have four important strucutural traits that facilitate the mechanism of catalysis described above.
-
Catalytic triad. It is a group of three side chains: aspartate (Asp), histine (His), and serine (Ser), which are close to each other in the active site of the protein, although they are far apart in the aminoacid sequence of the polipeptide chain. They work toghether: the histine (His) is a general base, that can accept the proton from the hydroxyl group of the reactive serine (Ser), in this way, this facilitates the formation of the covalent tethaedral transition state. The aspartate (Asp) stabizes the positive charge of histine (His).
-
Oxyanion hole. The stabilization and the tight binding of the tethaedral transition states is performed by providing a set of groups that forms hydrogen bonds to the negatively charged oxygen atom attached to the C1. These groups forms a compacted space, or pocket named the oxyanion hole. The positive charge that develops on the (histine) His residue after it has accepted a proton, also stabilizes the negative charged transition state.
-
Main chain substrate binding. In general, the serine protreinases have not substrate specifity. Thus, they can cleave peptide bonds with a variety of side chains adjacent to the scissible peptide bond. The cause is that polypetide substrates do not exhibit a specific binding to the enzyme through their main chain atoms, which form hydrogen bonds in a short antiparallel B sheet with main-chain atoms of a loop region in the enzyme.
-
Specifity pocket. As it has been stated below, the serine proteinases have not absolute specifity. Even so, many of them show a preferece for particular side chain before the scissile bond was seen from the amino end of the polypeptide chain. For instance, the chymotrypsin has preference to cleave after large aromatic side chains, and the trypsin has a marked preference to cleave after Lys, or Arg side chains. In each case, the preferred side chain is oriented so as to fit into a space, or pocket of the enzyme, typically named specifity pocket.
