The family S1 of Clan PA is subdivided in different subfamilies,that are phylogenetically distinct but share a common two beta-barrel architecture.
The S1A proteases subfamily are the trypsins that mediate a variety of extracellular processes. S1A proteases have a limited distribution in plants, prokaryotes, and the archaea. In humans, they are phylogenetically grouped into six functional categories: digestion, coagulation and immunity, tryptase, matriptase, kallikrein, and granzymes.
Activation of many trypsin-like serine proteases requires proteolytic processing of an inactive zymogen precursor. Cleavage of the proprotein precursor occurs at the identical position in all known members of the family, that is, between residues 15 and 16 (chymotrypsin numbering is conventionally used for sequence and structure comparisons). The nascent N-terminus induces conformational change in the enzyme through formation of an ion-pair with the highly conserved D194 that organizes both the oxyanion hole and substrate-binding site.
P98139|192-431|FVII_rabbit ---------------IVGGKVCPKGECPWQAALMNG--STLLCGGSLLDT 33
Q9GLP2|214-448|PC_pig ---------------LVNGKQSPWGESPWQVILLDSK-KKLACGAVLIHV 34
P00740|227-459|FIX_Homo ---------------VVGGEDAKPGQFPWQVVLNG-K-VDAFCGGSIVNE 33
P00743|234-466|FX_Bovis ---------------IVGGRDCAEGECPWQALLVNEE-NEGFCGGTILNE 34
P00735|367-621|Thrombin_Bos ---------------IVEGQDAEVGLSPWQVMLFRKSPQELLCGASLISD 35
sp|P00767|CTRB_BOVIN CGVPAIQPVLSGLARIVNGEDAVPGSWPWQVSLQDST-GFHFCGGSLISE 49
sp|P00761|TRYP_PIG --FPTDD-----DDKIVGGYTCAANSIPYQVSLNSGS---HFCGGSLINS 40
:* * . . *:*. * **. ::
The trypsin fold is remarkable for the even distribution of catalytic residues across the entire polypeptide sequence.Two six-stranded beta-barrels come together asymmetrically to host at their interface the residues of the catalytic triad H57 and D102 belong to the N-terminal beta-barrel with S195 and the oxyanion hole (S195 and G193) hosted by the C-terminal beta-barrel. Eight surface exposed loops in the protein decorate the entrance to the active site.
P98139|192-431|FVII_rabbit HWVVSAAHCFDKLSSLRNLTI-----VLGEHDLSEHE-GDEQVRHVAQLI 77
Q9GLP2|214-448|PC_pig SWVLTAAHCLDDY---KKLTV-----RLGEYDLRRRE-KWEVDLDIKEFL 75
P00740|227-459|FIX_Homo KWIVTAAHCVETG---VKITV-----VAGEHNIEETE-HTEQKRNVIRII 74
P00743|234-466|FX_Bovis FYVLTAAHCLHQA---KRFTV-----RVGDRNTEQEE-GNEMAHEVEMTV 75
P00735|367-621|Thrombin_Bos RWVLTAAHCLLYPPWDKNFTVDDLLVRIGKHSRTRYERKVEKISMLDKIY 85
sp|P00767|CTRB_BOVIN DWVVTAAHCGVTT-----SDV----VVAGEFDQGLET-EDTQVLKIGKVF 89
sp|P00761|TRYP_PIG QWVVSAAHCYKSR-----IQV----RLG-EHNIDVLE-GNEQFINAAKII 79
::::**** : . .
P98139|192-431|FVII_rabbit MPDKY--VPGKTDHDIALLRLLQPAALTNNVVPLCLPERNFSESTLATI- 124
Q9GLP2|214-448|PC_pig VHPNY--TRSTSDNDIALLRLAEPATFSQTIVPICLPDSGLSERELTRVG 123
P00740|227-459|FIX_Homo PHHNYNAAINKYNHDIALLELDEPLVLNSYVTPICIADKEYTN-IFLKF- 122
P00743|234-466|FX_Bovis KHSRF--VKETYDFDIAVLRLKTPIRFRRNVAPACLPEKDWAEATLMTQ- 122
P00735|367-621|Thrombin_Bos IHPRYN-WKENLDRDIALLKLKRPIELSDYIHPVCLPDKQTAAKLLHAG- 133
sp|P00767|CTRB_BOVIN KNPKFS--ILTVRNDITLLKLATPAQFSETVSAVCLPSADEDFPAGMLC- 136
sp|P00761|TRYP_PIG THPNFN--GNTLDNDIMLIKLSSPATLNSRVATVSLPRS--CAAAGTEC- 124
.: . ** ::.* * : : . .:.
P98139|192-431|FVII_rabbit PEVTGNMFCAGYLDGSK---DACKGDSGGPHATS--YHGTWYLTGVVSWG 213
Q9GLP2|214-448|PC_pig --ISENMLCAGILGDSR---DACEGDSGGPMVAS--FRGTWFLVGLVSWG 208
P00740|227-459|FIX_Homo --IYNNMFCAGFHEGGR---DSCQGDSGGPHVTE--VEGTSFLTGIISWG 206
P00743|234-466|FX_Bovis --ITPNMFCAGYDTQPE---DACQGDSGGPHVTR--FKDTYFVTGIVSWG 206
P00735|367-621|Thrombin_Bos --ITDNMFCAGYKPGEGKRGDACEGDSGGPFVMKSPYNNRWYQMGIVSWG 228
sp|P00767|CTRB_BOVIN --VTDVMICAG--ASGV---SSCMGDSGGPLVCQ--KNGAWTLAGIVSWG 216
sp|P00761|TRYP_PIG --ITGNMICVGFLEGGK---DSCQGDSGGPVVC----NG--QLQGIVSWG 202
: *:*.* .:* ****** . .. *::***
Within the same fold, trypsins prefer Arg/Lys side chains at the P1 position of substrate but chymotrypsins prefer Phe/Tyr/Trp residues. The molecular origin of this difference resides in part in the architecture of the primary specificity pocket, shaped as a cavity about 10 anstrom deep and connected to the active site, at the bottom of which trypsins carry D189 and chymotrypsins carry S189.
Also we find in trypsin family a conserved disulfide bridge pattern. Many serine protease domains contain four conserved disulfide bridges arranged in the following pattern: Cys 1-Cys 2; Cys 3-Cys 7; Cys 4-Cys 5; Cys 6-Cys 8.
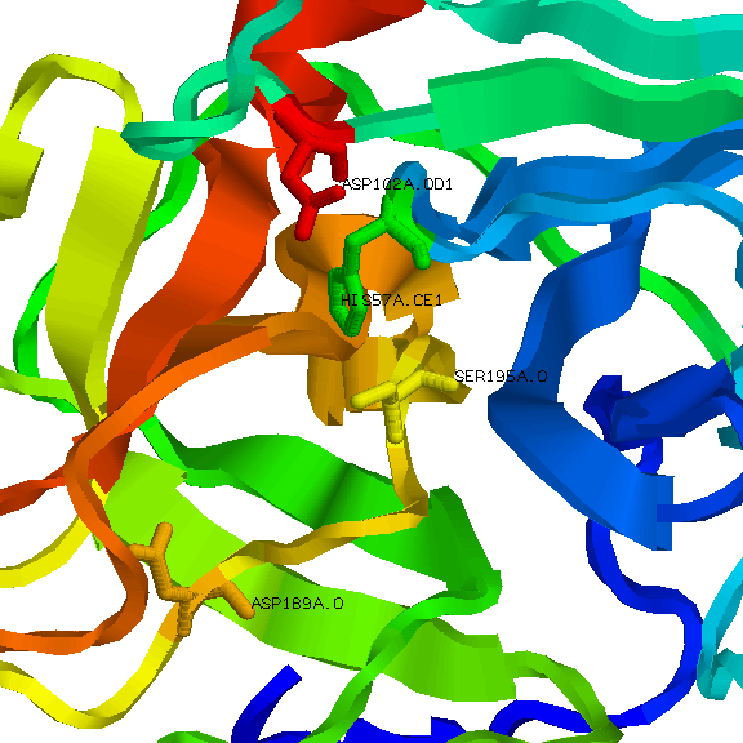
1.Trypsin cartoon representation 2.Trypsin active site