In biology, all the cascades and biochemical pathways have to be controlled and regulated in order to obtain and maintain a well-balanced system. In the context of this project, it is very important to state that blood coagulation is balanced by fibrinolysis process.
The important role of plasminogen in fibrinolyis makes it an interesting parameter to evaluate in various diseases. A decreased plasminogen level may in some situations compromise the body's ability to degrade fibrin and as such predispose to thrombosis. Hereditary plasminogen deficiency, as a cause of thrombosis, have also been reported in several cases. However, plasminogen deficiency is usually an acquired condition and since plasminogen is the inactive precursor of plasmin, most acquired defects are found in situations with increased fibrinolytic activity. An acquired deficiency is often seen with severe liver disease and acute disseminated intravascular coagulation (DIC), or as a result of thrombolytic therapy with plasminogen activators.
The blood coagulation process, as well as the fibrinolytic system, are heavily regulated and a whole set of inhibitors is present in the bloodstream for an efficient control and regulation. Even though this project is focused on coagulation process, it is interesting to present a brief description about the key component of the fibrinolytic system: plasminongen, as the the center of the process which works as a counterbalance of blood coagulation.
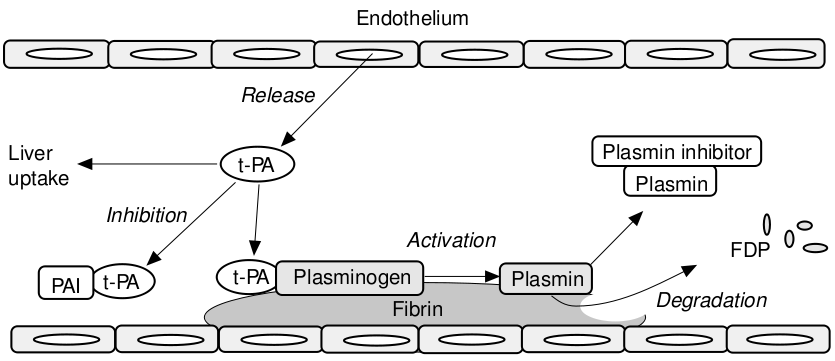
The central component in the fibrinolytic system is the glycoprotein plasminogen, which is produced by the liver and is present in plasma and most extravascular fluids. Plasminogen is a precursor enzyme (zymogen) which, following partial cleavage by a plasminogen activator is converted to its active and proteolytic form: plasmin. Plasmin is a serine proteinase which belongs to trypsin-like family and has a preferential cleavage of Lys, of Arg bonds. Therefore, its primary target is fibrin, but it is also able to degrade several constituents of the extracellular matrix and to convert a number of pro-hormones and cytokine precursors to their active form.
The generation of plasmin occurs preferentially on the fibrin surface, which offers binding sites for plasminogen and its principle activator in blood, t-PA. This binding stimulates plasminogen activation, but also localizes the action of plasmin to sites of fibrin formation which promotes efficient clotlysis. Further regulation is provided by the presence in plasma of inhibitors, primarily the plasmin inhibitor and the plasminogen activator inhibitor 1 (PAI-1).
The following picture is a schematic representation of the fibrinolyis mechanism. Plasminogen is the proenzyme of plasmin, whose primary target is the degradation of fibrin in the vasculature. t-PA (tissue-type plasminogen activator) is the principle activator of plasminogen in blood, while u-PA (urokinase plasminogen activator) is the predominant activator outside the bloodstream in the extracellular matrix. t-PA is produced by the vascular endothelial cells and is released into the circulation after stimulation. Fibrin regulates its own destruction by providing receptors or binding sites for plasminogen and t-PA, thus localizing the action of plasmin.
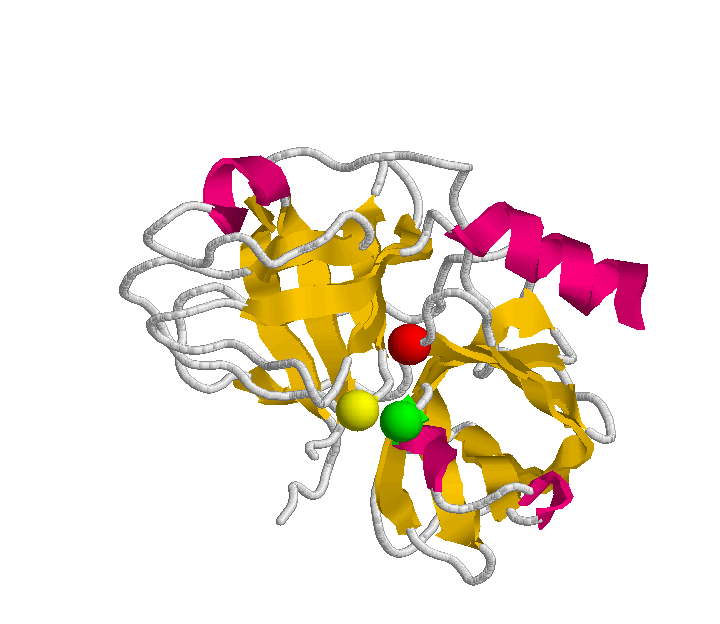
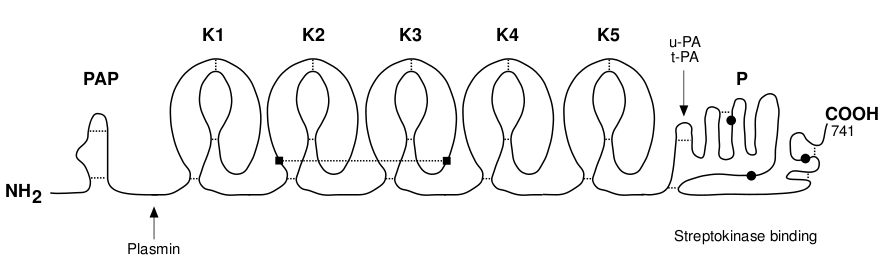
Human plasminogen is a single-chain glycoprotein containing 810 amino acid residues (P00747). The plasminogen molecule contains a total of six structural domains, each with different properties. The N-terminal portion of the molecule consists of five kringle domains with the capacity to bind to fibrin. The following figure deptics the primary structure of the plasminogen, that is the aminoacid sequence. A user-friendly color schema has been designed to provide an easy mechanism to detect all the region-domains, and the active site. This last it is composed by the typical catalytic triad of serine proteinases: Histidine (H), Aspartic acid (D), and Serine (S).
MEHKEVVLLL LLFLKSGQG
The following picture depcits a very basic schema of the plasminogen structure. It will be useful to make a comparision with the practical study done in the following sections. The plasminogen molecule consists of a set of several domains: preactivation peptide (PAN), five kringle domains and a trypsin-like domain.
As it can be seen above, plasminogen contains three different domains:
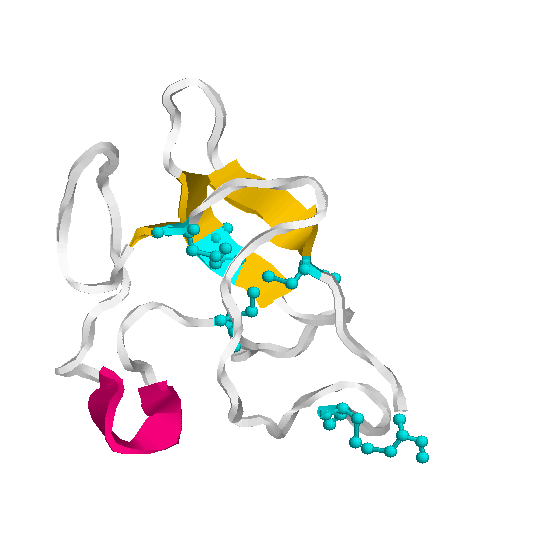
The kringle structure is characterised by a Cys cluster consisting of the two inner disulfide bridges (Cys 2 - Cys 4 and Cys 3 - Cys 5) that are almost perpendicular to each other and a terminal disulfide bridge Cys 1 - Cys 6. The binding of the ligand induces little conformational change in kringle 2 but stabilises its conformation.
The heavy chain of plasmin contains five kringle domains that carry lysine binding sites (LBS) which are responsible for an efficient binding to the substrate fibrin and to the inhibitor a2-antiplasmin and the light chain comprises the serine protease part with a partial phosphorylation site at Ser 578. These modules appear to function as independent domains.
Plasmin also acts as a proteolytic factor in many other physiological processes like wound healing, tissue remodelling, angiogenesis, embryogenesis and pathogen and tumor cell invasion. The main physiological inhibitor of plasmin is a2- antiplasmin.