This section presents an ortohology practical study, with the thrombin (bos taurus) as the center of the case study. To do this, two subsections are showed. In the first one, a primary structure comparision is performed. In the second subsection a structural comparision is presented. The main goal is to find similarities between the thrombin (bos taurus) and other thrombin proteins of other species.
The first step is to perform a multiple sequence alignment (MSA) of different thrombin proteins of a set of different species. With the aim to provide a progressive comparision, the MSA has been split in two series. In the first serie, a set of theoretical nearest species have been choosen (mammals). On the other hand, in the second serie a set of more distant species from bos taurus have been selected.
The following table contains a set of species close to bos taurus (in evolutionary terms):
| Uniprot Code | Name | Organism | Sequence length |
|---|---|---|---|
| P00735 | Prothrombin | Bos Taurus | 625 AA |
| P00734 | Prothrombin | Homo sapiens | 622 AA |
| P19221 | Prothrombin | Mus musculus | 618 AA |
| P18292 | Prothrombin | Rattus norvegicus | 617 AA |
| Q19AZ8 | Prothrombin | Sus scrofa | 623 AA |
| C8BKD1 | Prothrombin | Ovis Aries | 623 AA |
| Q5R537 | Prothrombin | Pongo Abelii | 623 AA |
ClustalW is the tool used to perform the MSA, and the results have been splitted in several pictures. Each one of this screenshots focuses in a specific part of the sequence.
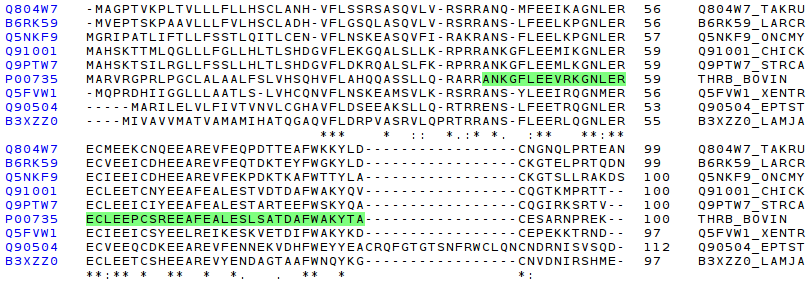
The most characteristic zone of serine-proteinases is the active site, composed of three aminoacids which creates the catalytic triad. In the first screenshoot these three aminoacids are red marked. The screenshot shows that the active (H, S, D) site is conserved along all the species.

Then, having seen the perfect conservation of active site in all thrombin proteins compared, it is very interesting to check if described protein domains of thrombin (bos taurus). The following picture shows a specific comparision of Gla, Krindle 1, and Krindle 2 domains.

It is easy to appreciate that the regions domains are very well conserved.
The last region to compare is the trypsin-like domain. This zone is thought to be evolved from an ancestral digestive protein (as it has been explain in coagulation evolution section). This infers a great region conservation of this protein region, as can be seen in the picture:
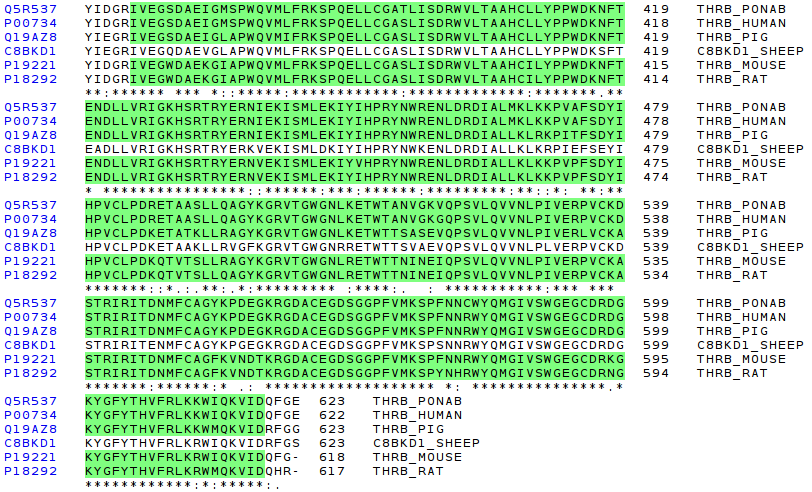
Following, a second MSA is presented. In this case, a set of further species from bos taurus (in evolution terms) have been selected. The objective is to check if the conservation trend observed in the MSA of above is observed there, with a set of species like fishes, or birds.
| Uniprot Code | Name | Organism | Sequence length |
|---|---|---|---|
| P00735 | Prothrombin | Bos Taurus | 625 AA |
| B6RK59 | Coagulation factor II | Larimichthys crocea | 618 AA |
| Q5NKF9 | Prothrombin | Oncorhynchus mykiss | 618 AA |
| Q91001 | Prothrombin | Oncorhynchus mykiss | 622 AA |
| Q9PTW7 | Prothrombin | Struthio camelus | 608 AA |
| Q5FVW1 | Coagulation factor 2 | Xenopus tropicalis | 607 AA |
| Q90504 | Thrombin | Eptatretus stoutii | 628 AA |
| B3XZZ0 | Coagulation factor 2 | Lampetra japonica | 605 AA |
| Q90504 | Thrombin | Eptatretus stoutii | 628 AA |
| Q804W7 | Prothrombin | Fugu Rubripes | 612 AA |
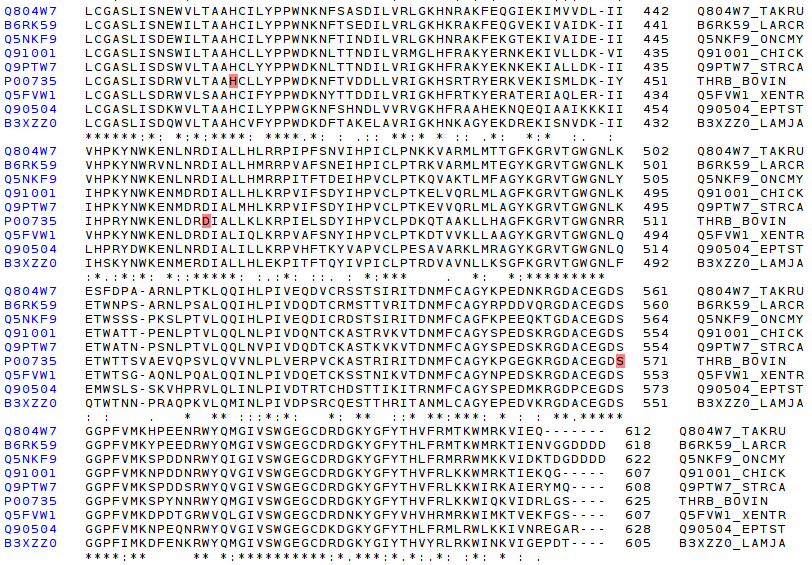
The first step is to check the catalitic triad, the active site of serine proteinases, which consists of three aminoacids (H, D, S). As the following picture shows, the catalytic triad is perfect conserved along all compared species.
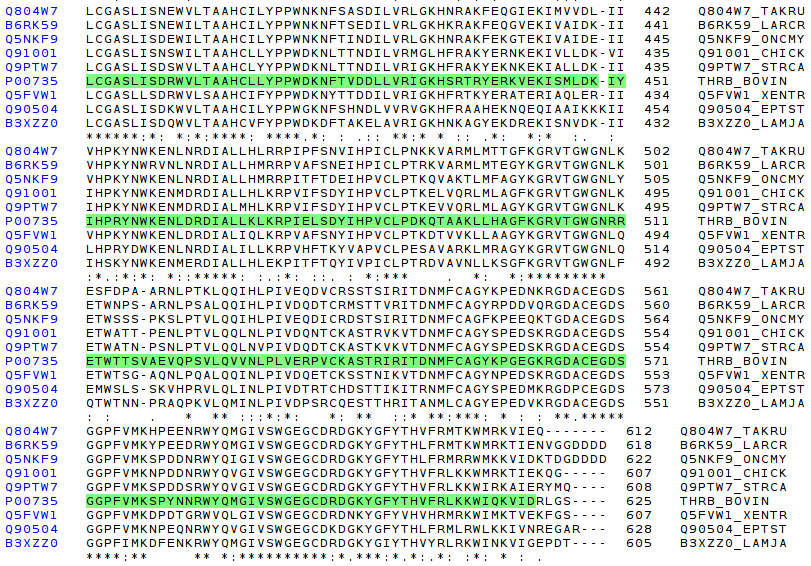
The following picture shows the Gla domain comparision, and the conserved region. It is highly conseverved, but less than in the first MSA serie of above.
The following pictures show the comparision of Kri1, and Kri2 domains. As it has happened in Gla domain, it is possible to notice a high conservation, but not so high like the first MSA.


Finally, the most conserved domain is trypsin like.
Having seen differences, and similarities at primary structure organization (multiple aminoacid sequence comparision), it is very worth to try to superpose different 3D structure (defined by X-ray techiniques) of thrombin proteins of different species. Unfortunately, only a few set of structures have been crystallographied.
This subsection presents two structural comparisions. Firstly, the following picture shows a structural superposition (done by STAMP) of the thrombin protein of bos taurus, and the thrombin of homo sapiens.
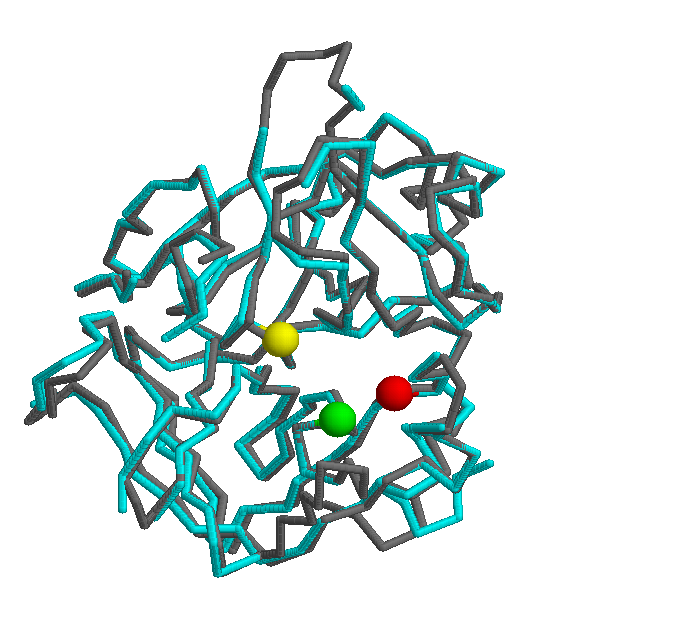
Another interesting comparision is against the mus musculus thrombin protein. The following picture shows a strucutural superpostion of these two orthologs.
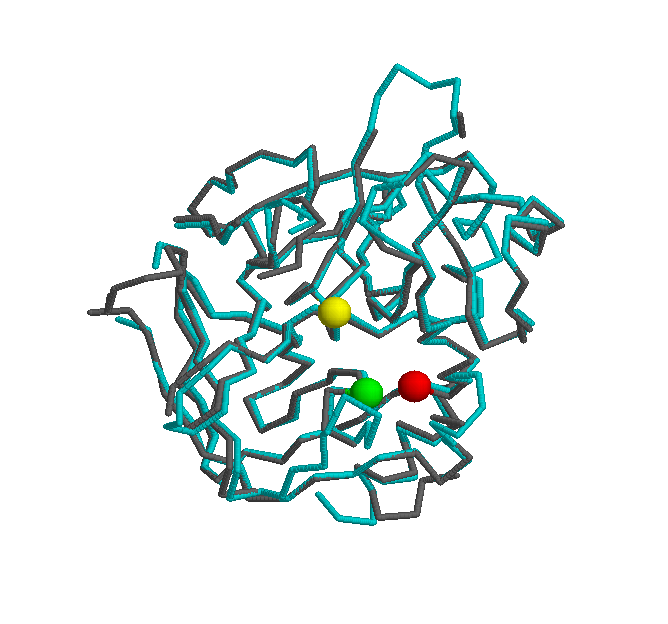
In the same way of the first structural superpostion (Bos taurus vs Homo sapiens), the thrombin structure is highly conserved. The active site (H - green, S - yellow, D - red) of both proteins shares the same postion, yielding a big similarity in the core of the protein.